Abstract
Background
The earliest records in Britain for the western European house mouse (Mus musculus domesticus) date from the Late Bronze Age. The arrival of this commensal species in Britain is thought to be related to human transport and trade with continental Europe. In order to study this arrival, we collected a total of 16 ancient mouse mandibulae from four early British archaeological sites, ranging from the Late Bronze Age to the Roman period.
Results
From these, we obtained ancient mitochondrial DNA (mtDNA) house mouse sequences from eight house mice from two of the sites dating from the Late Bronze to Middle Iron Age. We also obtained five ancient mtDNA wood mouse (Apodemus spp.) sequences from all four sites. The ancient house mouse sequences found in this study were from haplogroups E (N = 6) and D (N = 2). Modern British house mouse mtDNA sequences are primarily characterised by haplogroups E and F and, much less commonly, haplogroup D.
Conclusions
The presence of haplogroups D and E in our samples and the dating of the archaeological sites provide evidence of an early house mouse colonisation that may relate to Late Bronze Age/Iron Age trade and/or human expansion. Our results confirm the hypothesis, based on zooarchaeological evidence and modern mtDNA predictions, that house mice, with haplogroups D and E, were established in Britain by the Iron Age and, in the case of haplogroup E, possibly as early as the Late Bronze Age.
Similar content being viewed by others
Background
The western European house mouse (Mus musculus domesticus) is today a widely distributed commensal species that is closely associated with human settlements. Although likely a commensal since Neolithic times in the Near East, with rare evidence for its presence in Europe during the Bronze Age [1, 2], it has been argued, from zooarchaeological analyses, that it probably did not spread widely into western Mediterranean and northern Europe until the first millennium BC, during the Late Bronze Age or Iron Age [3]. The earliest records in Britain have been thought to date from the Late Bronze Age [4,5,6,7]. However, as these are not from securely dated stratigraphic contexts [8], it is possible that the house mouse did not arrive and become fully established Britain until the emergence of the denser human settlements of the Iron Age [3]. This may have initially taken place in southern England [9, 10], where the presence of structures to store cereal grain, which represent an ideal niche for house mice, would have helped their introduction, although cereals were available from the Neolithic [11]. While identification of their physical remains is not always straightforward, genetic evidence can give clearer results.
Within the last 3000 years, the British Isles have experienced multiple waves of human immigration and a long history of contact, trade and exchange with continental Europe. Due to the close relationship between the two species, demographic changes in human populations are likely paralleled by similar changes in house mouse numbers; indeed, the house mouse niche has largely been shaped by humans [12]. Indeed, phylogeographic studies have shown that historical human movements have impacted on current house mouse population patterns [7, 12,13,14,15]. However, to date, the founding populations and colonisation routes have been identified based on modern phylogeography alone. Ancient DNA (aDNA) analyses of early house mice from Great Britain can test these conclusions and help us to understand the origins of the earliest house mouse populations in the British Isles.
In this study, we aim to characterise the first colonisation of the western European house mouse in Great Britain, and the possible route that these first arrivals followed, based on ancient DNA. To this end, we analysed mitochondrial DNA (mtDNA) control-region sequences of some of the rare ancient house mouse remains from four archaeological sites in southern England, dating from the Late Bronze Age to the Middle Iron Age and Roman period, and compared these with published modern European mtDNA control-region data.
Methods
Archaeological samples
The archaeological record of small rodents is adversely affected by their small size, and sieving of sediment using a maximum mesh of 2 mm is necessary for their recovery [11, 16]. The small size of the bones also makes them problematic for radiocarbon dating, as a single bone will rarely yield enough collagen for direct analysis. Therefore, the dating of mouse material often requires dating by context and association with artefacts and, consequently, lacks reliability and precision.
We collected a total of 16 ancient mouse mandibulae from four British archaeological sites, ranging from the Late Bronze Age to the Roman period (see Additional File 1: Fig. S1; Table S1)—Potterne, Wiltshire (n = 7); Battlesbury Bowl, Wiltshire (n = 5); North West Farm, Dorset (n = 2); and Druce Farm Roman Villa, Dorset (n = 2).
The site of Potterne, near Devizes, Wiltshire, was excavated by the commercial archaeological unit Wessex Archaeology between 1982 and 1984, and comprises an extensive accumulation of dark anthropogenic soil deposits, up to 2 m deep in places, covering an area of 3.5 ha. The ‘midden-like’ deposits were rich in artefacts and biological remains, resulting from the accumulation of manure and refuse from stock keeping, and the repeated dumping and trampling of waste from human occupation and activities on and around the site over a 500-year period. Pottery typology, and radiocarbon dating of charcoal from different levels within the deposit and other cut features, suggest a date of 1200–600 BC, encompassing the Late Bronze Age into the very Early Iron Age period [6]. In addition to a large animal bone assemblage dominated by domestic mammals, small mammal remains were recovered, mainly from sieved environmental samples, and house mouse remains were identified from every level [17]. Although it was not possible to obtain direct radiocarbon dates from the mice themselves to confirm the Late Bronze Age date, a radiocarbon date of 1,460–990 cal. BC (2ơ; lab number HAR-8938) was obtained from charcoal that came from the same posthole as mouse mandible OG06 [6]. The assumption is that all seven mouse samples (OG01–OG07) are contemporaneous with the associated archaeological materials of the Late Bronze Age (c. 1200–600 cal. BC) and date from the same layers and contexts; although we note that Locker [17] does caution that some small mammal remains may have filtered down the deposit from higher levels.
The later prehistoric site of Battlesbury Bowl lies along a narrow chalk ridge immediately to the north of Battlesbury Camp, an Iron Age hillfort near Warminster, Wiltshire. Excavations by Wessex Archaeology in 1999 revealed features of Late Bronze Age to Middle Iron Age dates (based on ceramic style), including ditches, post holes, and almost 200 pits [18]. The faunal assemblage is one of the largest collections of Early to Middle Iron Age faunal material from Great Britain. Hambleton and Maltby [19] report the presence of both house mouse and wood mouse in both the hand-recovered assemblage and the environmental sieved samples. The mouse mandibulae included in this study came from pit fills (OG08, OG09, OG11, OG12) and a posthole (OG10), all of which were assigned Early to Middle Iron Age dates. Radiocarbon dating of a pig humerus, from the same context as mouse mandible OG11, provided a date of 420–100 cal. BC (2ơ; lab number NZ-13634) [18].
The site at North West Farm is just outside the village of Winterborne Kingston to the north of Bere Regis, Dorset. The archaeology represents multiple phases spanning Bronze Age to Roman periods. It forms part of a program of archaeological fieldwork, the Durotriges Project, designed to investigate native and Romano-British settlement across Dorset, focusing specifically on the archaeologically distinct Iron Age Durotriges tribe. Two mouse remains (OG13 and OG14) were recovered from a chalk deposit (340) within one of three large storage pits in Trench H of the 2017 fieldwork season and, based on preliminary pottery attributions and the form of the pits, these may date to the Bronze Age.
Druce Farm Villa, Puddletown, Dorset, comprises a series of stone and flint constructed, and timber-post built, buildings arranged on a courtyard plan, surrounded by a series of ditched enclosures with features associated with industrial use (e.g. kilns/ovens and pits) [20]. The site displays a number of phases of use in Romano-British times between the first and fourth centuries AD. Two mouse samples (OG15 and OG16) were obtained from an extensive deposit of remains of microfauna, which lay on the intact mosaic floor of a room in the main range of buildings, sealed by a deposit of degraded plaster and roof tiles. Analysis of the site and the deposit are ongoing [20], but this appears to represent a deposit of owl pellets, most likely derived from barn owls, which accumulated when the building was going out of use, and was sealed by the collapsed roof. The mosaic floor has been typologically dated to the fourth century AD. Two water vole mandibles from the deposit were successfully subjected to radiocarbon dating (which was possible because this is a relatively large rodent that can yield enough collagen for AMS radiocarbon dating) to elucidate the date of the building collapse, and returned dates of cal. AD 249–391 and cal. AD 208–346 (both 2ơ; [20]).
Morphological identification
The morphological identification of our 16 mouse mandibles was not easily resolved, as the characters published to distinguish house mouse (Mus spp.) from Apodemus spp. [21, 22] are not always applicable. Furthermore, some of the archaeological specimens were either missing the M1 tooth (OG05 and OG16), where the distinguishing characters are present or visible, or the M1 was in an advanced wear stage (OG04, OG05, OG07, OG08 and OG09).
The character we used to determine species identification was the presence of tubercles on the buccal side of the M1 in Apodemus spp., which are absent in Mus spp. [21]. Some of the specimens analysed were suspected to be Apodemus spp. but were sampled for DNA to see if the presence of tubercles could be a reliable diagnostic trait (Table 1).
Extraction and amplification of ancient DNA
We undertook sample processing at the Ancient DNA Facility of the University of Huddersfield (UK) under dedicated clean-room conditions supplied by a positive air pressure system. Researchers wore full body suits, hairnets, gloves and face masks throughout the sampling, extraction and PCR set-up processes, and constantly cleaned all tools and surfaces with LookOut® DNA Erase (SIGMA Life Sciences), as well as with bleach, ethanol and exposures to UV light.
We decontaminated the surface of the mandibulae by UV radiation for 10 min on each side. We shook whole or partial jaws with a zirconium oxide grinding ball inside a zirconium oxide jar in a Mixer Mill (Retsch MM400) for 15 s at 30 Hz/s. We extracted DNA from the resulting 10–50 mg of powder produced following the protocol by Yang et al. [23], with modifications by MacHugh et al. [24]. We included blank controls throughout the sampling procedure, extraction, and PCR set-up to monitor for possible contamination.
We amplified and sequenced the mtDNA sequences in 12 overlapping 121–150 bp fragments (Table 2) covering a 915-bp fragment of the control region. We designed four primer pairs specifically for this study, took six from Jones et al. [25] with minor modifications, and designed new reverse primers for two primer sets (see Table 2 for more detail). Each primer pair amplified overlapping fragments, including the most variable region between positions 15,381 and 15,663 (when compared to the reference Mus musculus domesticus mitochondrial genome, accession number NC_006914).
We designed three of the pairs of primers (2b, 2c and 3) that can also amplify the DNA of Apodemus. This was done to help identify mandibulae that presented identification difficulties where identification as Mus was uncertain.
Analyses of ancient and modern house mouse data
Phylogenetic reconstruction from Mus control-region data is challenging, generating even more homoplasy than is seen in modern humans. We estimated phylogenies in two different ways. We firstly aligned the eight ancient sequences (OG01, OG02, OG03, OG04, OG07, OG08, OG09 and OG10) obtained for Mus musculus domesticus in this study with 728 previously published house mouse sequences from Austria, Bulgaria, Great Britain, Croatia, Denmark, France, Germany, Greece, Italy, Ireland, Norway, Portugal, Spain and Sweden (see Additional File 1: Table S2) [7, 25,26,27,28, 31, 33,34,35,36] to create a Bayesian inference phylogenetic tree with MrBayes [29], using the parameters previously calculated in JModelTest [30]. We ran the analysis for five million generations with four chains and with a 25% burn-in. We used FigTree v.1.3.1 to visualise the tree, and haplogroups were assigned and named following previous nomenclature [26].
The necessity of using shorter sequence lengths in the aDNA analysis meant that it is not possible to identify all of the clades revealed in the original published house mouse analyses (that used longer sequence lengths). Therefore, we used these data to estimate haplogroup phylogenies using the Network software (see Additional File 1: Table S3), constructing separate networks for lineages from haplogroups D and E using Network v.5.0.1.1 (www.fluxus-engineering.com). After an initial run to separate the major haplogroups (not shown), we first made the data binary where necessary (at positions with both a transition and a transversion, or a transition and a deletion), and then ran the reduced-median algorithm, followed by the median-joining algorithm, on the pre-processed dataset. We included indels, as a proportion of the variation in the mouse control region comprises insertion events; however, where there were tracts of contiguous indels (such as the 11 base pair insertion seen in some D1 individuals between positions 16,089 and 16,090 when compared to the reference mitogenome, NC_006914), we only counted these as a single event. We estimated the position of the root of each haplogroup network from the larger network.
Results
Sample identification
In total, we obtained mtDNA sequence data from 13 of the 16 samples, of which we identified eight individuals as Mus musculus domesticus, four as Apodemus sylvaticus, and one as Apodemus flavicolis (Table 2). The Bronze Age North West Farm site only yielded DNA from one individual, attributed to A. sylvaticus, and the Roman period Druce Farm site also yielded only A. sylvaticus (n = 2). Other Apodemus spp. samples were found at the Late Bronze Age/Early Iron Age site of Potterne (A. sylvaticus; n = 1) and the Iron Age Battlesbury site (A. flavicolis; n = 1). However, in both of these sites, eight M. m. domesticus samples were also found (five at Potterne and three at Battlesbury). These results highlight the uncertainty in the identification of murid species in the archaeological record based on mandible morphology, particularly if the M1 is either worn or absent. We tested the reliability of the presence of tubercles on the buccal side of the M1 being a diagnostic trait for Apodemus spp., as these are absent in Mus spp. [21]. In one case, OG03, the M1 was present, and unworn tubercles appeared also to be present, suggesting that the specimen belonged to Apodemus. However, the specimen was partially obscured by sediment, and the mtDNA indicated the specimen to be Mus spp. (Table 2). In all other cases, the diagnostic nature of the tubercles proved reliable, and aDNA was shown to be a useful tool for species identification if wear or a lack of M1 did not allow the trait to be used.
House mouse phylogeography in Britain
The longest fragment obtained from M. m. domesticus was 772 bp (OG04), from positions 15,508 to 16,279 of the reference house mouse mitogenome (NC_006914; Table 2). The shortest fragment (OG01) had a length of only 74 bp. All house mouse individuals analysed here date to the Late Bronze Age or the Iron Age, and they clustered within two main haplogroups (D and E; Table 3; Fig. 1), described previously in modern samples from Britain.
Bayesian phylogenetic tree constructed from 772 bp of house mouse control region sequences (from positions 15,508 to 16,279 of the reference house mouse mitogenome, NC_006914). The tree includes 728 modern and eight ancient British sequences (Additional File 1: Table S2). Ancient samples are indicated with a black dot. Bootstrap values of greater than 50 are indicated on the branches
Six individuals (from across both sites) belonged to haplogroup E and two more (from Battlesbury Bowl) clustered in haplogroup D. Haplogroup F, the most widespread cluster in Britain today, was not present in our sample set. Three samples (OG04, OG07 and OG08) belonged to the same haplotype within E.
The haplogroup networks (Fig. 2) locate the ancient British samples within haplogroups D and E. All five Potterne samples and the Battlesbury Bowl haplogroup E sequence are securely located in close proximity to French/British/Irish samples within haplogroup E. Four of them directly match the root haplogroup of this group, found both in modern Britain and France, which is related to lineages seen in Potterne, modern Great Britain and Ireland. The two remaining Battlesbury Bowl samples fall within haplogroup D. Haplogroup D is itself rare in present-day Britain and confined to the north in current datasets [26, 31].
Phylogenetic networks for haplogroups D and E, constructed from 772 bp of house mouse control region sequences (from positions 15,508 to 16,279 of the reference house mouse mitogenome, NC_006914). The modern sequences used to construct each network are shown in Additional File 1: Table S3. Haplogroup D network includes two of the ancient house mouse sequences (both from Battlesbury Bowl), while the haplogroup E network includes six of the ancient house mouse sequences (five Potterne and one Battlesbury Bowl). The ancient samples include OG01, 03, 09 and 10, with the asterisk in the haplogroup E network indicating OG02, 04, 07 and 08. Where there are missing data in the ancient sequences (Table 3), the sample has been placed determined on its most likely position based on the phylogenetic network. The precise position of OG10 within the D/D1 clade in the phylogenetic network may be unreliable due to missing data. Circles represent sequence haplotypes, the area being proportional to the frequency, and they are coloured by the location of the samples, as seen in the accompanying key. Small grey points are reconstructed intermediate nodes introduced by the network algorithm, and links between haplotypes represent mutations. R denotes the likely position of the root of each network, as determined from the full network (not shown)
Unfortunately, the fragments obtained for the Apodemus spp., whilst distinctive, were too short for meaningful integration into wood mouse (A. sylvaticus) or yellow-necked mouse (A. flavicolis) phylogeographies.
Discussion
If M. m. domesticus started its spread from the Near East, associated with modern humans, during the Early Neolithic [3, 37, 38], then Britain is at the periphery of the western European expansion of the subspecies. The phylogeography of the western part of the Atlantic geographical range of the house mouse has been particularly well studied [7, 26, 39] and, based on haplotype diversity and the presence of different clades, Britain has a relatively high genetic variation [7]. This study complements the understanding of the colonisation of the British Isles by the western European house mouse by showing that two haplogroups, D and E, were present in southern Great Britain by the time of the Iron Age, although haplogroup E may have arrived earlier based on its presence on the Late Bronze Age/Early Iron Age archaeological site (Fig. 3). These two clades may represent several different mouse migrations linked to different human movements.

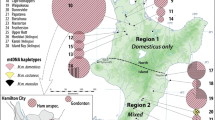
adapted from Mapbox (www.mapbox.com)
Localities and mtDNA haplogroups for all samples of Mus musculus domesticus, the western European house mouse, from the British Isles and northern France. Colours indicate the control region haplogroup presence with several individuals (white: haplogroup C; orange: haplogroup D; red: haplogroup E; blue: haplogroup F). Branches are not proportional to the number of mutations. Circles, squares, stars and triangles show previously published sequence data as denoted in the key. Crosses show the localities where ancient DNA has been recovered as part of this study. Credit: Image
Haplogroup E is the most common M. m. domesticus lineage in modern southern and central British samples and is also present in Scotland and southern Ireland (Figs. 1, 2 and 3) [7, 26]. Searle et al. [7] suggested that the distribution of haplogroup E might reflect the colonisation of Great Britain from the European mainland during the Iron Age. As this clade is not well represented in central Europe, it has been suggested that they did not arrive with people overland but via a maritime route, possibly transported from the Mediterranean by the seafaring Phoenicians in the Late Bronze Age/Early Iron Age [32]. The network of E, even with the truncated sequences, (Fig. 2) could point toward a putative proximal source in Germany for the French, British and Irish lineages, with several distinct subclades, most of which Great Britain shares with France, and a suggestion of a possible ancestry further back in Portugal, and ultimately Greece, as previously published [32]. Although zooarchaeological evidence has been used to argue that there was little or no house mouse presence in western Europe until the urban developments of the Iron Age [3], our results complement the possibility that the extant haplogroup E lineages may have reached southern Great Britain as early as the Late Bronze Age.
Haplogroup D is much less common in Britain today (Fig. 3). It is distributed at low frequencies from the Levant to the central and western Mediterranean, and north into Germany, northern Britain and the Baltic. Its derived subclade, D1, co-occurs with the remainder of D in Germany, but has replaced it in Denmark and Sweden (but not Norway), where D1 reaches 100%. D1 is also seen in Madeira and the Canary Islands [31, 33, 39], and the colonisation of these islands has been attributed to Danish Viking movements, first to Madeira and then to the Canary Islands, following Portuguese settlement [31, 39]. Only a single instance of D1 has been recorded in the British Isles, from the far north, and haplogroup D overall is restricted to Scotland in current published data.
The presence of haplogroup D in Britain in the Iron Age, and more particularly in southern England, may suggest an introduction from continental Europe—either Germany or Denmark, or possibly even Iberia. These lineages are phylogenetically distinct from extant British D lineages, which appear more directly linked to lineages from Germany, and may be unconnected to them historically (Fig. 2). Haplogroup D may, therefore, have been introduced to Britain on at least two distinct occasions, all of them possibly separate from the introduction of haplogroup E (Fig. 3).
Although three main haplogroups are represented in modern British samples (D, E and F, with F being the most frequent; Fig. 3), only two of these (D and E) have been found in the archaeological sites analysed here. The presence of haplogroups D and E in Britain since at least the Iron Age had previously been hypothesised from the analysis of modern samples [25, 26, 32]. This study provides the first direct evidence of the presence of D and E in Britain, and specifically southern England, from at least the Iron Age period, and the lack of haplogroup F in our data is consistent with its later introduction, possibly associated with the Vikings [7].
Conclusion
The presence of haplogroups D and E during the Late Bronze Age and Iron Age in Great Britain has provided evidence of an early house mouse colonisation that may be related with first-millennium expansions of humans. This is broadly in agreement with what has been previously suggested on the basis of modern mtDNA data, although the presence of haplogroup E possibly in the Late Bronze Age would predate, by several centuries, the most widely accepted model for its expansion in Europe [32, 38].
Availability of data and materials
All the ancient British house mouse mtDNA sequences generated in the course of the study have been deposited in GenBank (Accession Numbers MW459973-MW459980). Public access to the database is open.
References
Brothwell DR. The Pleistocene and Holocene archaeology of the house mouse and related species. Symp Zool Soc Lond. 1981;47:1–13.
Ijzereef GF. Bronze Age animal bones from Bovenkarspel; the excavation at Het UzlkJe. Amersfoort. 1981; (Nederlandse Oudheden, 10).
Cucchi T, Vigne JD, Auffray JC. First occurrence of the house mouse (Mus musculus domesticus Schwarz & Schwarz, 1943) in the western Mediterranean: a zooarchaeological revision of subfossil occurrences. Biol J Lin Soc. 2005;84(3):429–45.
Brady R, Ellison A. Ram’s Hill: a Bronze Age defended enclosure and its landscape. BAR Brit Ser. 1975;19:1–9.
Bell M. Brean Down excavations 1983–87. In: Engl. Heritage Archaeol. Rep. 15. London, UK: Historic Buildings and Monuments Commission for England; 1990.
Lawson AJ. Potterne 1982–85. Animal husbandry in later prehistoric Wiltshire. In: Wessex Archaeol. Rep. 17; 2000.
Searle JB, Jones CS, Gündüz İ, Scascitelli M, Jones EP, Herman JS, Rambau RV, Noble LR, Berry RJ, Giménez MD, Jóhannesdóttir F. Of mice and (Viking?) men: phylogeography of British and Irish house mice. Proc R Soc B. 2009;201:56.
O’Connor T. Extinctions and Invasions. In: O’Connor TP, Sykes N, editors. A social history of British fauna. Oxford: Oxbow Books; 2010. p. 127–33.
Coy J. The bird bones in Cunliffe, B W Danebury: An Iron Age Hilfort in Hampshire. Vol. 2: The excavations 1969–78: The finds. London. In: CBA Research Report 52(2), 527–531; 1984.
Harcourt R. The animal bones in Wainwright, G J Gussage all Saints: An Iron Age Settlement in Dorset. London. In: Department of Environment Archaeological Report 10:150–160; 1979.
Oconnor T. Economic prehistory or environmental archaeology? On gaining a sense of identity. In: Albarella U, editor. Environmental archaeology: meaning and purpose environmental science and technology library. Dordrecht: Springer; 2001.
Gabriel SI, Stevens MI, Mathias MDL, Searle JB. Of mice and ‘convicts’: Origin of the Australian house mouse, Mus musculus. PLoS ONE. 2011;6(12):e28622.
García-Rodríguez O, Andreou D, Herman JS, Mitsainas GP, Searle JB, Bonhomme F, Hadjisterkotis E, Schutkowski H, Stafford R, Stewart JR, Hardouin EA. Cyprus as an ancient hub for house mice and humans. J Biogeogr. 2018;45:2619–30.
Hardouin EA, Chapuis JL, Stevens MI, Van Vuuren JB, Quillfeldt P, Scavetta RJ, Teschke M, Tautz D. House mouse colonization patterns on the sub-antarctic Kerguelen archipelago suggest singular primary invasions and resilience against re-invasion. BMC Evol Biol. 2010;10:45.
Lippens C, Estoup A, Hima MK, Loiseau A, Tatard C, Dalecky A, Bâ K, Kane M, Diallo M, Sow A, Niang Y, Piry S, Berthier K, Leblois R, Duplantier JM, Brouat C. Genetic structure and invasion history of the house mouse (Mus musculus domesticus) in Senegal, West Africa: a legacy of colonial and contemporary times. Heredity. 2017;119:64.
O’Connor TP, Barrett JH. Animal bones. In: Balme J, Paterson A, editors. Archaeology in practice: a student guide to archaeological analyses. Oxford: Blackwell; 2006. p. 260–95.
Locker A. Animal bone. In: Lawson, A.J. Potterne 1982–5: Animal Husbandry in Later Prehistoric Wiltshire. Salisbury: Wessex Archaeology Report 17:101–19; 2000.
Ellis C, Powell AB. An Iron Age Settlement Outside Battlesbury Hillfort, Warminster and Sites Along the Southern Range Road. Salisbury, England. Salisbury: Wessex Archaeology; 2008.
Hambleton E, Maltby M. Faunal remains. In: An Iron Age Settlement Outside Battlesbury Hillfort, Warminster and Sites Along the Southern Range Road. Salisbury, England, C. Ellis & A. B. Powell. Salisbury: Wessex Archaeology, 84–93; 2008.
Ladle L, pers. comm
Chaline J, Baudvin H, Jammot D. Saint-Girons MC. Les proies des rapaces (petits mammifères et leur environnement). Paris: Doin; 1974.
Hilson S. Teeth. Cambridge: Cambridge University Press; 1986.
Yang DY, Eng B, Waye JS, Dudar JC, Saunders SR. Technical note: improved DNA extraction from ancient bones using silica-based spin columns. Am J Phys Anthropol. 1998;105:539–43.
MacHugh DE, Edwards CJ, Bailey JF, Bancroft DR, Bradley DG. The extraction and analysis of ancient DNA from bone and teeth: a survey of current methodologies. Anc Biomol. 2000;3:81–102.
Jones EP, Skirnisson K, Mcgovern T, Gilbert M, Willerslev E, Searle JB. Fellow travellers: a concordance of colonization patterns between mice and men in the North Atlantic region. BMC Evol Biol. 2012;12(1):1–8.
Jones EP, Jóhannesdóttir F, Gündüz İ, Richards MB, Searle JB. The expansion of the house mouse into north-western Europe. J Zool. 2011;283(4):257–68.
Nachman MW, Boyer SN, Searle JB, Aquadro CF. Mitochondrial DNA variation and the evolution of Robertsonian chromosomal races of house mice Mus domesticus. Genetics. 1994;136(3):1105–20.
Prager EM, Sage RD, Gyllensten ULF, Thomas WK, Hübner R, Jones CS, Noble LES, Searle JB, Wilson AC. Mitochondrial DNA sequence diversity and the colonization of Scandinavia by house mice from East Holstein. Biol J Lin Soc. 1993;50(2):85–122.
Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. Mrbayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–42.
Posada D. Jmodeltest: phylogenetic model averaging. Mol Biol Evol. 2008;25(7):1253–6.
Bonhomme F, Orth A, Cucchi T, Rajabi-Maham H, Catalan J, Boursot P, Auffray JC, Britton-Davidian J. Genetic differentiation of the house mouse around the Mediterranean basin: Matrilineal footprints of early and late colonization. Proc R Soc B. 2011;278(1708):1034–43.
Bonhomme F, Searle JB. House mouse phylogeography. In: Macholán M, Baird SJE, Munclinger P, Piálek J, editors. Evolution of the house mouse. New York: Cambridge University Press; 2012. p. 278–98.
Gündüz İ, Auffray JC, Britton-Davidian J, Catalan J, Ganem G, Ramalhinho MG, Mathias ML, Searle JB. Molecular studies on the colonization of the Madeiran archipelago by house mice. Mol Ecol. 2001;10(8):2023–9.
Ihle S, Ravaoarimanana I, Thomas M, Tautz D. An analysis of signatures of selective sweeps in natural populations of the house mouse. Mol Biol Evol. 2006;23:790–7.
Prager EM, Tichy H, Sage RD. Mitochondrial DNA sequence variation in the eastern house mouse, Mus musculus: comparison with other house mice and report of a 75-bp tandem repeat. Genetics. 1996;143:427–46.
Rajabi-Maham H, Orth A, Bonhomme F. Phylogeography and postglacial expansion of Mus musculus domesticus inferred from mitochondrial DNA coalescent, from Iran to Europe. Mol Ecol. 2008;17:627–41.
Hardouin EA, Orth A, Teschke M, Darvish J, Tautz D, Bonhomme F. Eurasian house mouse (Mus musculus L.) differentiation at microsatellite loci identifies the Iranian plateau as a phylogeographic hotspot. BMC Evol Biol. 2015;15(1):1–12.
Cucchi T, Papayianni K, Cersoy S, et al. Tracking the Near Eastern origins and European dispersal of the western house mouse. Sci Rep. 2020;10:8276.
Förster DW, Gündüz İ, Nunes AC, Gabriel S, Ramalhinho MG, Mathias ML, Britton-Davidian J, Searle JB. Molecular insights into the colonization and chromosomal diversification of Madeiran house mice. Mol Ecol. 2009;18(21):4477–94.
Acknowledgements
The authors would like to thank the Wiltshire Museum for the zooarchaeological material and Lilian Ladle for information about Druce Farm. The authors also would like to thank Maria Pala and Jarek Bryk for commenting on an earlier version of the manuscript, and the three anonymous reviewers who gave useful feedback.
Funding
No funding was obtained for this study.
Author information
Authors and Affiliations
Contributions
OG-R, CJE, MBR and JRS wrote the manuscript. OG-R and CJE completed the lab work. OG-R, CJE and EAH undertook data analyses. EH, JM and CR provided information about the zooarchaeological context and the material. JRS and OG-R undertook the identification of the material. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The material from Potterne and Battlesbury Bowl was sampled with permission from Lisa Brown, the Curator of the Wiltshire Museum in Devizes. The material from Druce Farm and North West Farm were sampled with permission from two of the authors who excavated and worked on the sites (JM and CR).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Supplementary material including Table S1, Table S2, Table S3 and Figure S1.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
García-Rodríguez, O., Hardouin, E.A., Hambleton, E. et al. Ancient mitochondrial DNA connects house mice in the British Isles to trade across Europe over three millennia. BMC Ecol Evo 21, 9 (2021). https://doi.org/10.1186/s12862-021-01746-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12862-021-01746-4