Abstract
Background
Hypoxic culture conditions have been used to study the impact of oxygen deprivation has on gene expression in a number of disease models. However, hypoxia response elements present in the promoter regions of some commonly used housekeeping genes, such as GAPDH and PGK1, can confound the relative gene expression analysis. Thus, there is ongoing debate as to which housekeeping gene is appropriate for studies investigating hypoxia-induced cell responses. Specifically, there is still contradicting information for which housekeeping genes are stable in hypoxia cultures of mesenchymal stem cells. In this study, candidate housekeeping genes curated from the literature were matched to RNAseq data of normoxic and hypoxic human adipose-derived stem cell cultures to determine if gene expression was modulated by hypoxia or not. Expression levels of selected candidates were used to calculate coefficient of variation. Then, accounting for the mean coefficient of variation, and normalised log twofold change, genes were ranked and shortlisted, before validating with qRT-PCR. Housekeeping gene suitability were then determined using GeNorm, NormFinder, BestKeeper, comparative\(\Delta Ct\), RefFinder, and the Livak method.
Results
Gene expression levels of 78 candidate genes identified in the literature were analysed in the RNAseq dataset generated from hADSC cultured under Nx and Hx conditions. From the dataset, 15 candidates with coefficient of variation ≤ 0.15 were identified, where differential expression analysis results further shortlisted 8 genes with least variation in expression levels. The top 4 housekeeping gene candidates, ALAS1, RRP1, GUSB, and POLR2B, were chosen for qRT-PCR validation. Additionally, 18S, a ribosomal RNA commonly used as housekeeping gene but not detected in the RNAseq method, was added to the list of housekeeping gene candidates to validate. From qRT-PCR results, 18S and RRP1 were determined to be stably expressed in cells cultured under hypoxic conditions.
Conclusions
We have demonstrated that 18S and RRP1 are suitable housekeeping genes for use in hypoxia studies with human adipose-derived stem cell and should be used in combination. Additionally, these data shown that the commonly used GAPDH and PGK1 are not suitable housekeeping genes for investigations into the effect of hypoxia in human adipose-derived stem cell.
Similar content being viewed by others
Background
Housekeeping genes (HKG) are stable, constitutively expressed reference genes which are used to normalise the expression of genes-of-interest in a variety of molecular techniques. HKG need to be ubiquitously and stably expressed in order to accurately quantify any treatment-related differential gene expression. For this purpose, it is usual to select HKG involved in supporting cellular functions [1]. Typically, HKG are selected from related literature, or most commonly determined from previous work performed in the same laboratory, sometimes without sufficient regard to the stability of that HKG to the different treatments being applied. The stability of HKG expression should be examined for each experimental design, and specific cell types [2], as it cannot be assumed that a “one-size-fits-all” approach is suitable.
Hypoxic (Hx) culture conditions have been used to model cellular physiological responses in various ischemic disorders, such as ischemic brain injury [3, 4], tumour progression and hypoxic microenvironment [5, 6], diabetes and obesity [7, 8], and toxin-induced reactive oxygen species in chronic kidney diseases [9]. The molecular response to hypoxia is typically driven by hypoxia response elements (HRE) present in the promoter region of genes. Functional HRE contain a target cis-acting DNA sequence that binds hypoxia-inducible factor -1 (HIF-1) protein complex which regulates gene expression [10, 11]. As hypoxia is a fundamental change to culture conditions, other genes regulated by hypoxia can be involved in metabolic switching pathways such as redox homeostasis and glycolysis, hence a large number of complex interactions make it difficult to select for HKG based on the absence of HRE alone. A number of commonly used HKG contain HRE and are regulated by hypoxia, for example GAPDH [12,13,14] and PGK1 [10]. A list of 114 genes which are induced by hypoxia is shown in Additional file 1 [15, 16]).
While various HKG have been reported as suitable for hypoxia studies (summarised in Table 1), there is ongoing debate in the literature as to which HKG are stably expressed in hypoxia [17,18,19,20,21,22,23,24,25,26,27], especially in mesenchymal stem cells [20, 28,29,30]. Thus, using a standard protocol for identifying multiple reference genes, we selected RPL13A, TBP and GAPDH from the literature and performed a preliminary qRT-PCR to study gene regulation in Hx human adipose-derived stem cells (hADSC), but found that these genes, unlike what they’ve been reported, were not stably expressed (Additional file 2). This represents a major setback which requires an alternative solution.
Alternative to testing potential HKG from the literature one at a time, approaches have been developed to identify and quantify a panel of selected HKG candidates based on the gene expression values from qRT-PCR data, such as GeNorm [31] which uses a geometric averaging method, NormFinder [32] a model-based variance estimation method, BestKeeper [33] a pair-wise comparison method, comparative \(\Delta Ct\) method [34] by relative expression, and RefFinder [35] which uses a geometrically averaging weighted ranks for the listed methods above. However, these approaches require numerous qRT-PCR experiments with a set of at least 12 HKG candidates and involve a larger number of samples and reagents.
With the advancement of next generation sequencing technology, RNA sequencing (RNAseq) is an attractive tool that can be used to obtain a broad comparison between transcriptome profiles [1]. Some recent studies have used RNAseq datasets to select stable HKG [36,37,38] customized for a unique experimental design. Lemma et al. 2018 [37] selected HKG from RNAseq data using normalised log2 fold change (L2FC), while Tang et al. 2017 [36] and Wang et al. 2019 [38] normalised for coefficient of variation of mRNA transcript expressions when selecting for HKG. Due to the complex nature of gene expression and regulation by hypoxia, RNAseq may be an effective way to screen large transcriptome data for HKG selection.
In this study, we aim to use RNAseq data from hADSC to screen HKG from the literature and validate their expressions under hypoxic regulation, in order to identify the most suitable HKG in Hx hADSC. RNAseq datasets were generated from hADSC from 5 donors and cultured under Nx or Hx conditions in vitro. RNAseq and bioinformatic analysis was used to identify differences in the transcriptome between donors and culture conditions. Specifically, the stability of candidate HKG in cells cultured under Nx and Hx conditions was determined by calculating the coefficient of variation (CV) of both normalised and transformed read counts, and L2FC obtained from differential gene expression. Potential HKG candidates were identified by low CV (≤ 0.15), ranked and were validated using qRT-PCR where relative expression of each selected HKG was calculated using the \({2}^{-\Delta \Delta Ct}\) Livak method [39], GeNorm method, NormFinder method, the BestKeeper method, \(\Delta Ct\) method and RefFinder method.
Results
Differential gene expression analysis
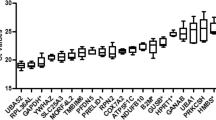
To identify which genes were differentially expressed by hADSC cultured under Hx conditions, the RNAseq transcriptional profile of five primary hADSC cell cultures (hADSC-A, -B, -C, -D, -E) grown under Hx conditions were compared to the profile of hADSC cells cultured under standard conditions (Nx). Analysis of the transcriptome revealed a total of 2800 differentially expressed transcripts; Fig. 1 shows the top 50 transcripts of at least |2| L2FC. DESeq2 was also used to cluster each dataset according to the similarities in transcript count distribution (Fig. 2). Principal Component Analysis (PCA, Fig. 3) identified distinct clusters of differentially expressed genes in hADSC cultured in Nx and Hx conditions, as well as a donor-to-donor variability in transcriptome profile between the hADSC lines.
Principal Component Analysis (PCA) of hADSC datasets. PCA plot of hADSC lines (A-E, colour-coded) in hypoxic (Hx, triangles) and normoxic (Nx, circles) treatment groups. The first principal component (PC1) represents the maximum variance direction of data distribution at 52%, and the second principal component (PC2) represents the second largest variation at 24%
Comparison of hypoxic effects on HKG identified from the literature
Candidate HKG were identified from the literature (ACTB, ALAS1, B2M, GAPDH, GUSB, HPRT1, PGK1, POLR2B, PPIA, RPL13A, RPLPO, RRP1, TBP, TFRC and YWHAZ) and matched for normalised gene expression in the RNAseq dataset generated from hADSCs cultured under Hx and Nx conditions (Fig. 4). Normalised counts indicated a similar distribution between Hx and Nx datasets for 7 of the candidates (ACTB, ALAS1, B2M, GUSB, POLR2B, RRP1 and TBP). In contrast, another 7 genes (GAPDH, HPRT1, PGK1, PPIA, RPL13A, RPLP0 and YWHAZ) increased expression in Hx samples compared to Nx hADSC, and TFRC showed reduced expression in Hx hADSC.
Selection of HKG from the RNAseq dataset
To compare the levels of gene expression, data was first expressed in transcripts per million (TPM) normalised counts and DESeq2-normalised counts for the selected HKG. Both TPM and DESeq2-normalised counts were then used to calculate for CV. before the list of HKG were ranked smallest to largest for both CV and L2FC (Additional file 3). Then, the average CV was calculated from HKG with CV ≤ 0.15, before normalising CV and L2FC to the smallest corresponding values of the 78 candidates and summed. HKG were ranked from smallest to the largest value for the sum of these four methods. Table 2 shows the top 8 (least variation) HKG candidates calculated and the top 4 listed in Table 3 for validation with qRT-PCR, with data analysis workflow summarised in Additional file 4.
Selection for low CV in the Hx hADSC found 8 of 78 HKG candidates screened had a \(\underline{CV}\) ≤ 15%. The top 4 HKG were validated further by qRT-PCR along with PGK1 and 18S mRNA expression detected in four hADSC cell lines cultured under Nx and Hx conditions. Table 3 lists the raw Ct values for each HKG (ALAS1, RRP1, 18S, PGK1 and GUSB) in each hADSC (POLR2B was not detected in all hADSC but was expressed in a control cell line (keratinocytes) at raw Ct = 25.0 ± 0.92). Of the expressed HKG, GUSB showed very high Ct values in hADSC-A, -B and -D, indicating very low expression of the gene (keratinocytes showed expression at raw Ct = 25.5 ± 0.08). Hence, both POLR2B and GUSB were unsuitable HKG for hADSC in hypoxia studies due to the low to no expression levels on qRT-PCR.
Ct values were validated using GeNorm (geometric average), NormFinder (model-based variance estimation), BestKeeper (pair-wise correlations), comparative delta-Ct, RefFinder (geometric average of weighted ranking) and Livak (\({2}^{-\Delta \Delta Ct}\)) methods to quantify the stability of HKG expression in hADSC cultured under Nx and Hx conditions. Figure 5a-e summarises the ranking of the 5 HKG, where the closer each value is to 1, the more stably expressed each HKG were. Figure 5f shows the relative expression (\({2}^{-\Delta \Delta Ct}\)) of each gene using one of the HKG as a reference gene at each time. Based on the combined analysis of all 6 methods (Fig. 5g), 18S and RRP1 were identified as most stable of the 5 HKG tested in our culture conditions.
Comparative study of published methods for quantitating variance in normalised Ct values of housekeeping genes in hADSC under Nx and Hx culture conditions. Raw Ct values of the 5 HKG in hADSC under Nx and Hx conditions were normalised and ranked using A) GeNorm (geometric mean), B) NormFinder (model-based variance estimation), C) BestKeeper (pair-wise correlations) D) comparative delta-Ct, E) RefFinder (geometric mean of each ranking), F) relative expression calculated with 2−△△Ct (Livak and Schmittgen 2001) using all HKG candidates as reference genes in a step-wise manner to compare between Hx and Nx, and G) The final ranking of all HKG candidates in each hADSC
Further validation of the suitability of 18S and RRP1 as HKG (Fig. 6) was performed. The expression of a known hypoxia-regulated gene – VEGFA, was normalised to 18S and RRP1 and the levels of expression in cells treated with Hx compared to those cultured under Nx conditions. The relative expression of VEGFA was stable across 2 biological replicates of hADSC (hADSC-C and -D) cultured in hypoxia or normoxia. Figure 6 shows calculated relative expression (\({2}^{-\Delta \Delta Ct}\)) of VEGFA was up-regulated 6.8-fold ± 1.4 when using 18S as HKG, compared to a 12-fold ± 0.6 upregulation when using RRP1 as HKG. Used in combination, using the geometric mean of the two selected HKG, a 9.5-fold ± 1.0 upregulation in VEGFA gene expression was measured.
Validation of 18S and RRP1 housekeeping genes by qRT-PCR analysis for hypoxic stimulation of VEGFA expression. Relative expression of VEGFA in hypoxic versus normoxic treated hADSC (\({2}^{-\Delta \Delta Ct}\)). Relative expression was calculated using the geometric mean of both 18S and RRP1 reference genes (N = 3 replicates, experiment repeated twice. Data are represented as mean ± SD)
Discussion
Since some typical HKG were found to be regulated in hADSC by hypoxia, this study used RNAseq datasets to screen the transcriptome for suitable HKG candidates to validate differential gene expression levels, regulated by hypoxia, by qRT-PCR. Further validation was performed with qRT-PCR and analysed using 6 established methods to calculate the variation in relative gene expression. 18S and RRP1 were identified as stable HKG suitable for use in hypoxia studies in hADSC in vitro. The suitability of HKG candidates was determined by having low variation in the gene expression levels across test and control samples. Furthermore, the raw Ct values of 18S and RRP1 fell within the Ct range expected of high copy number and commonly used HKG genes, which usually range between 14 to 32 raw Ct [40] (Ct values are inverse proportional to gene expression levels). In addition, the Ct of these candidate HKG were no more than 10 raw Ct values apart from the Ct of the genes-of-interest which is important to reduce variation as a result of amplification errors. qRT-PCR validation identified the following candidate HKGs suitable for hypoxia studies in hADSC, these genes have been ranked from the most stable to least stable: 1) 18S, 2) RRP1, 3) ALAS1, 4) PGK1 and 5) GUSB.
Comparison between the RNAseq-screening approach and qRT-PCR (the gold standard), identified only RRP1 and PGK1 as suitable HKG for hypoxia studies. The result for GUSB, ALAS1 and POLR2B were not consistent between these methods. 18S, an rRNA not identified in the transcriptome data since the library preparation was enriched for mRNA, was validated by qRT-PCR against VEGFA, a known hypoxia-regulated gene, was one of the most stable genes tested in this study, evident also in a previous study [17]. This represents a limitation in using RNAseq to identify for HKG as this approach does not account for total RNA and may exclude potential non-mRNA HKG candidates.
One criterion for HKG candidates was to be constitutively expressed in normal cell functions [1]. 18S rRNA is a small subunit rRNA, which together with the large subunit rRNA forms ribosomes essential for mRNA translation. RRP1, encodes for the nuclear protein ribosomal RNA processing protein 1 homolog A, which is recruited at late stages of nucleologenesis during mitosis [41]. From the validation experiments using VEGFA as a known hypoxia-regulated gene, both 18S and RRP1 fulfilled the selection of HKG based on cellular functions, and stable replicable relative expressions with VEGFA in 2 biological hADSC replicates (hADSC-C and -D).
Both 18S and RRP1 are suitable HKG for hADSC hypoxia studies either individually, or used in combination. 18S is expressed at a higher level (Ct ~ 20) and RRP1 has a lower expression (Ct ~ 30), which allows users the option to select the HKG expressed at a similar level to their gene-of-interest to not overestimate relative gene expression analysis. Alternatively, according to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines [28, 42], it is recommended to use a combination of multiple HKG using geometric mean.
Apart from stable HKG, the results also identified genes that are not suitable as HKG for hADSC hypoxia studies. ALAS1 is a mitochondrial enzyme where cellular respiration and cell response to hypoxia takes place [43]. It was found to be significantly down-regulated (2−△△Ct ≤ -1.4-fold change) when normalised to 18S and hence not suitable HKG for hADSC hypoxia studies. PGK1, encoding for a glycolytic enzyme involved in glucose metabolism and a known regulator of hypoxia, was confirmed as a Hx-regulated gene in hADSC in both RNAseq and qRT-PCR results, acting as a positive control measure in gene regulation under hypoxia. GUSB, ranked less stable than PGK1, encodes for beta-glucuronidase, which is an enzyme located in the lysosomes involved in glycosaminoglycan degradation. GUSB was expressed at high Ct values, indicating low gene expression, suggesting a down-regulation of GUSB and the production of beta-glucuronidase as a response to hypoxia regulation and hence not an appropriate HKG for this study.
In addition to the genes validated in qRT-PCR, other commonly used HKG were also shown to be significantly regulated by hypoxia in hADSC (adj-P ≤ 0.05) transcriptome data. These included GAPDH (23.12 = 8.7-fold), HPRT (20.53 = 1.4-fold), PGK1 (23.53 = 11.6-fold), PPIA (20.78 = 1.7-fold), RPL13A (21.23 = 2.3-fold), RPLP0 (21.58 = 3.0-fold), YWHAZ (20.39 = 8.7-fold) and TFRC (2–3.23 = -9.4-fold). The literature indicated that HRE were identified in the promoter regions of PGK1 [10], GAPDH [13] and TFRC [44], but not HPRT, PPIA, RPL13A, RPLP0 or YWHAZ, suggesting an indirect hypoxia response to these genes. Hence, this study provides added information to exclude genes that were unsuitable as HKG for Hx hADSC, in the absence of HRE in corresponding promoter regions.
In comparison to the only other study in the literature comparing HKG in hADSC cultured under hypoxic conditions [20], this study identified different findings for 18S and GUSB being suitable HKG, but also similar findings in identifying unsuitable HKG such as GAPDH. While it is accepted that GAPDH is regulated by hypoxia and not suitable to be used as a HKG, the difference in findings for 18S and GUSB might be contributed to hADSC being cultured in slightly different growth conditions in Fink et al., such as high versus low passages, and hypoxia cultures over 1 and 2 weeks. Our culture conditions have been optimised for the experimental question, and hence reinforcing the need to validate suitable HKG according to experimental design.
Limitations of study
RNA sequencing was performed in two separate batches and the differential expression analysis did not correct for batch effects. However, the selection of HKG using RNAseq data for CV calculation was independently performed between the hADSC lines. As the second sequencing experiment (for hADSC-A, -B, -C and -D) was sequenced more in-depth (25 to 57 million reads) than the first (for hADSC-E at 5.9 to 8.7 million reads), more confidence was given if CV ≤ 0.15 was more consistent in hADSC-A, -B, -C and -D when selecting for HKG after the calculations.
This study described a straightforward RNAseq-based screening approach to identify potential HKG, with the assumption that normalised count reads represent gene expressions. However, the TPM normalisation method may underestimate genes that are expressed at very low levels. For example, hADSC-C mean expression for CDKN1A was 163.5 TPM ± 19.1 TPM at CV 11.7%, but for CHFR was 0.6 ± 0.1 TPM at CV = 23.1%. In this scenario, CDKN1A would be defined as “stable expression” and included for subsequent analysis, but CHFR, where there was only 0.1 TPM standard deviation of count reads, would be excluded. This may be resolved if the selection for potential HKG is performed for genes expressed at similar levels as specific gene-of-interests of the study.
Additionally, this study may be limited by specificity of probe/primer designs in qRT-PCR, as POLR2B was expressed in both RNAseq normalised counts, but when used for qRT-PCR validation, were found to not be expressed at detectable levels. Further evaluation of qRT-PCR primers for POLR2B isoforms in hADSC may be warranted in future studies.
Conclusions
In conclusion, this study found RNAseq data able to identify candidate housekeeping genes for use in hypoxia studies of human adipose-derived mesenchymal stem cells. After validation by qRT-PCR, 18S rRNA and RRP1 were identified as suitable HKG candidates and are recommended as stable housekeeping genes for future hypoxia-induced gene expression studies in hADSC. The differential expression of several commonly used housekeeping genes, provided further evidence to eliminate them as suitable for HKG in future hypoxia studies of human adipose-derived stem cells.
Materials and methods
Cell culture maintenance
hADSC from 5 donors (A, B, C, D and E) were purchased from Lonza (Lonza, Walkersville, Maryland) or ZenBio (ZenBio, Research Triangle Park, North Carolina), where cells were previously characterised (general information on hADSC donors and cell characterisation are provided in Additional file 5). hADSC were cultured in Dulbecco’s Modified Eagle’s medium (DMEM) containing 1 g/L D-glucose (Gibco, Carlsbad, California), 2 mM GlutaMAX™ (hADSC-E in 4 mM L-glutamine) and 110 mg/L Sodium Pyruvate, supplemented with 10% foetal bovine serum (FBS, Bovogen, Keilor East, Australia) and 1% Penicillin–Streptomycin (P/S; Gibco, Carlsbad, California). Spent media were replaced every 2–3 days. Cells were incubated in standard culture conditions of 37 °C, with 5% carbon dioxide (CO2) and 21% atmospheric oxygen (O2) in a humidified cell culture incubator unless otherwise specified, and passaged at approximately 80% confluence. hADSC were used between passage 5 to 8.
A human eardrum keratinocyte cell line characterised previously [45] was cultured in DMEM containing 4.5 g/L D-glucose (Gibco, Carlsbad, California), 4 mM L-glutamine) and 110 mg/L Sodium Pyruvate, supplemented with 10% FBS and 1% P/S. Spent media were replaced every 3–4 days. Cells were incubated in standard culture conditions of 37 °C, with 5% CO2 and 21% atmospheric oxygen (O2) in a humidified cell culture incubator and passaged at approximately 90% confluence. Keratinocytes of passage 36 were used.
Hypoxia conditioning
hADSC were cultured to 80–90% confluence in 75 cm2 tissue culture flasks, rinsed with phosphate-buffered saline (PBS), pH 7.2 (Gibco, Carlsbad, California), and cultured in serum-free DMEM for at least 8 h before replacing with fresh serum-free medium for a further 48 h. hADSC at this step were either incubated in standard (Nx; 37 °C, 5% CO2) or Hx conditions in the GENbox Jar (BioMérieux, Craponne, France) produced by an anaerobic atmosphere generator sachet (BioMérieux, Craponne, France) at 37 °C. A hypoxia indicator strip (BioMérieux, Craponne, France) was included in the sealed chamber to confirm gas composition (< 0.1% O2 and 15% CO2). At the end of the 48-h incubation, total RNA was extracted.
RNA extraction
Total RNA from hADSC-E was extracted using the FavorPrep™ Tissue total RNA Mini Kit (Favorgen Biotech Corp, Ping Tung, Taiwan) according to the manufacturer’s protocol, before eluted RNA was cleaned up of genomic DNA (gDNA) using the RNeasy Plus Micro kit (Qiagen, Hilden, Germany). Total RNA from hADSC-A, -B, -C, -D, and keratinocytes were extracted using the RNeasy Plus Micro kit according to the manufacturer’s instructions. Briefly, cells were lysed and gDNA was removed via the gDNA Eliminator spin column. Total RNA was then bound, collected and washed on the RNeasy MinElute spin column before being eluted at approximately 14 μL per sample. All RNA samples were stored at -80 °C until required for RNA sequencing and cDNA synthesis.
RNA samples were submitted to the Australia Genome Research Facility (AGRF, Melbourne, Australia) for quality control, library preparation and RNA sequencing. Briefly, RNA samples were quantified using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, California) where samples of at least an RNA Integrity Number (RIN) of ≥ 8.0 and an A260/280 ratio of 1.8 – 2.0 were accepted. RNA quantification results are provided in Additional file 6.
Library preparation and RNA sequencing
For all samples, library preparation was performed using the Illumina TruSeq® Stranded mRNA kit according to the manufacturer’s protocol (Illumina, San Diego, California). Briefly, mRNA was purified and fragmented for double-stranded cDNA synthesized using specific methods to improve strand specificity. Adapters were ligated to DNA fragments to prepare for hybridization onto a flow cell during RNA sequencing, and enriched by amplification.
For hADSC-E, samples were processed earlier in a separate batch to the rest. RNA sequencing was performed on the Illumina HiSeq 2500 platform High Output Mode using the HiSeq Control Software (HCS) v2.2.68 and Real Time Analysis (RTA) v1.18.66.3 running on the Illumina sequencing computer for single-end reads at 50 bp. The Illumina bcl2fastq 2.18.0.12 pipeline was used next to generate the sequence data.
For hADSC-A, -B, -C and -D, RNA sequencing was performed on the Illumina NovaSeq 2500 platform High Output Mode using the NovaSeq Control Software (NCS) v 1.6.0 and Real Time Analysis (RTA) v 3.4.4 performing real-time based calling on the NovaSeq instrument computer for single-end reads at 100 bp. The Illumina bcl2fastq v 2.20.0.422 pipeline was used next to generate the sequence data. All RNAseq files were uploaded onto ArrayExpress accession E-MTAB-9979.
Bioinformatic analysis
RNAseq raw fastq files were checked using the FastQC (v 0.11.9) tool [46] to evaluate the quality of the sequenced samples. Reads were trimmed with Trimmomatic (v 0.36) [47] with parameters recommended in the Trimmomatic tool manual for single-end reads. Trimmed reads were then aligned to the GrCh38 human reference genome assembly obtained from Ensembl using the HISAT2 (v 2.1.0) tool [48], with the default parameters in the HISAT2 tool manual. Mapped reads were processed using SamBamba (v 0.6.6) [49]. For hADSC-A, -B, -C and -D, lane 1 files were merged with their corresponding repeated lane 2 files. Multimappers were filtered out using a mapping quality threshold of ≥ 5. Sorted and filtered Bam files were used as input for the featureCounts function in subread [50] to count each transcript to the GrCh38.90 reference genome. Sequencing details and processing steps are attached in Additional file 7. Count files (number of reads per transcript counted in data files) were parsed to include the corresponding Ensembl Gene IDs, before analysing with DESeq2 (v 1.26.0) [51] via R studio.
Differential expression analyses were performed using DESeq2 to compare differential gene expressions from hADSC cultured in Hx against Nx. Low count reads (< 10) were removed and DESeq2 was used with the multi-factor design for “conditions” (Hx vs Nx) and “cell types” (hADSC-A, -B, -C, -D and -E). Count data were transformed using regularised logarithmic transformation (rlog function) in DESeq2 to compare between datasets in the clustering analyses. Differential expression analysis was performed on raw count data and results were shrunk using the Apeglm function [52] for better estimation of distribution and exported as L2FC. Transcripts were considered differentially expressed (DE) if L2FC ≥ 2 at adjusted-P ≤ 0.05.
In addition to the L2FC data, raw count reads from all 5 hADSC were also normalised to transcripts per million (TPM) by normalising for both gene lengths and sequence depths.
All analysis performed in R for this study can be found in Additional file 4.
Ranking method
From the literature, a list of 78 previously known HKG were curated. L2FC and DESeq2-normalised values were extracted from the differential expression analysis data of the 5 hADSC, and TPM were calculated using the formula below, for the 78 HKG.
The CV of each HKG between Hx and Nx was calculated with TPM or DE data using the following formula below, for all 5 hADSC (CVABCDE) as well as individually (CVA, CVB, CVC, CVD, CVE):
An average of CV (\(\underline{CV}\)) was first calculated using only CV ≤ 0.15. For example, ALAS1 had 7 CV values ≤ 0.15, the average CV was calculated by averaging the 7 CV values only. Next, the top 8 genes were selected to normalise for both CV and L2FC by dividing each value with the smallest corresponding values for an arbitrary score. For example, from the averaged CV, ALAS1 was the smallest at 0.013 so average CV of all other genes were normalised to 0.013, and of the L2FC, GUSB was the smallest (closest to 0) at 0.01 so L2FC of all other genes were normalised to 0.01. These two arbitrary scores were then summed and ranked from the smallest to the largest where the top 4 HKG were selected for validation via qRT-PCR. Additional file 3 contains all 78 HKG data, calculations, and ranking.
cDNA synthesis
Complementary DNA was synthesised using the RT2 Omniscript kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. Briefly, 1 μg of template RNA was reverse transcribed in accordance with the manufacturer’s protocol for each sample, together with the corresponding no reverse transcriptase controls (NRT) and no template controls (NTC). For each reaction, 1 μM oligo(dT)15 primers (Promega Corporation, Fitchburg, Wisconsin) were used, where reactions were incubated at 37 °C for 60 min.
qRT-PCR
qRT-PCR was performed using the QuantiFast Probe PCR kit (Qiagen, Hilden, Germany) on cDNA generated from hADSC-A, -B, -C and -D RNA samples. Gene expressions were performed using FAM-labelled and VIC-labelled TaqMan Assay probes (Thermo Fisher Scientific, Waltham, Massachusetts) as listed below in Table 4. 18S was not included in our RNAseq since it’s an rRNA, VEGFA was to validate the known regulation by hypoxia. Briefly, 2.5 μL of cDNA, 5 μL of 2 × PCR Mastermix, 0.5 μL of TaqMan Assay probes and RNase-free H2O were combined to a total of 10 μL per reaction per well in a white 96-well full-skirt plate, along with appropriate controls for NRT and NTC. qRT-PCR was performed on the CFX Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, California) under the following conditions: 95 °C for 3 min, and 40 cycles of denaturation at 95 °C for 3 s and annealing at 60 °C for 30 s. Threshold cycles (Ct) were determined using the exponential growth phase and the baseline signal from fluorescence versus cycle number plots.
Validation of HKG stability
Using RefFinder [35], raw Ct values for each HKG of all 4 hADSC lines were entered according to the instructions to calculate for geometric average (GeNorm), model-based variance estimation (NormFinder), pair-wise correlations (BestKeeper), \(\Delta Ct\) method, as well as the geometric mean of the weighted ranks from the 4 methods to be expressed as RefFinder rank. Then, relative gene expression levels were determined using \({2}^{-\Delta \Delta Ct}\) method [39] for each hADSC to compare Hx (test) against Nx (control). The overall rank was averaged for each HKG. Relative gene expression levels (n = 3) using \({2}^{-\Delta \Delta Ct}\) was also performed for VEGFA against 18S and RRP1 in 2 biological replicates of hADSC in Hx (test) against Nx (control).
Availability of data and materials
The datasets generated during this study are available in the ArrayExpress repository with the accession code E-MTAB-9979. For correspondence and material requests, please address them to HTO.
Abbreviations
- qRT-PCR:
-
Real-time quantitative polymerase chain reaction
- cDNA:
-
Complementary DNA
- POLR2B:
-
RNA polymerase II subunit B
- ALAS1:
-
5’-Aminolevulinate synthase 1
- GUSB:
-
Glucuronidase beta
- RRP1:
-
Ribosomal RNA processing 1
- 18S:
-
18S Ribosomal RNA
- PGK1:
-
Phosphoglycerate Kinase 1
- VEGFA:
-
Vascular endothelial growth factor A
- Ct:
-
Threshold cycles
- NRT:
-
No reverse transcriptase control
- NTC:
-
No template control
- HKG:
-
Housekeeping gene
- TPM:
-
Transcript per million
- Hx:
-
Hypoxic
- Nx:
-
Normoxic
- L2FC:
-
Log 2 Fold Change
- CV:
-
Coefficient of Variance
- hADSC:
-
Human adipose-derived stem cells
- ACTB:
-
Beta-actin
- RPLP0:
-
Ribosomal protein lateral stalk subunit P0
- TFRC:
-
Transferrin receptor
- B2M:
-
Beta-2-microglobulin
- RPL13A:
-
Ribosomal protein L13a
- RPII:
-
RNA polymerase II
- 28S:
-
28S rRNA
- HPRT1:
-
Hypoxanthine phosphoribosyltransferase 1
- YWHAZ:
-
Tyrosine 3/tryptophan 5-monooxygenase activation protein
- TBP:
-
TATA-box binding protein
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- PPIA:
-
Peptidylprolyl isomerase A
- mRNA:
-
Messenger RNA
- HRE:
-
Hypoxia response elements
- HIF-1:
-
Hypoxia-inducible factor -1
References
Eisenberg E, Levanon EY. Human housekeeping genes, revisited. Trends Genet. 2013;29(10):569–74. https://doi.org/10.1016/j.tig.2013.05.010.
Caradec J, Sirab N, Keumeugni C, Moutereau S, Chimingqi M, Matar C, et al. ‘Desperate house genes’: the dramatic example of hypoxia. Br J Cancer. 2010;102:1037–43. https://doi.org/10.1038/sj.bjc.6605573.
Marti HH, Bernaudin M, Petit E, Bauer C. Neuroprotection and angiogenesis: dual role of erythropoietin in brain ischemia. Physiology News Physiol Sci. 2000;15:225–9. https://doi.org/10.1152/physiologyonline.2000.15.5.225.
Schmidt-Kastner R, Haberkamp M, Schmitz C, Hankeln T, Burmester T. Neuroglobin mRNA expression after transient global brain ischemia and prolonged hypoxia in cell culture. Brain Res. 2006;1103(1):173–80. https://doi.org/10.1016/j.brainres.2006.05.047.
Maxwell PH, Dachs GU, Gleadle JM, Nicholls LG, Harris AL, Stratford IJ, et al. Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proc Natl Acad Sci U S A. 1997;94(15):8104–9. https://doi.org/10.1073/pnas.94.15.8104.
Freese KE, Kokai L, Edwards RP, Philips BJ, Sheikh MA, Kelley J, et al. Adipose-derived stem cells and their role in human cancer development, growth, progression, and metastasis: a systematic review. Cancer Res. 2015;75(7):1161–8. https://doi.org/10.1158/0008-5472.can-14-2744.
Yin J, Gao Z, He Q, Zhou D, Guo Z, Ye J. Role of hypoxia in obesity-induced disorders of glucose and lipid metabolism in adipose tissue. Am J Physiol Endocrinol Metab. 2009;296(2):E333–42. https://doi.org/10.1152/ajpendo.90760.2008.
Hodson L, Humphreys SM, Karpe F, Frayn KN. Metabolic signatures of human adipose tissue hypoxia in obesity. Diabetes. 2013;62:1417–25. https://doi.org/10.2337/db12-1032.
Khanh VC, Ohneda K, Kato T, Yamashita T, Sato F, Tachi K, et al. Uremic toxins affect the imbalance of redox state and overexpression of prolyl hydroxylase 2 in human adipose tissue-derived mesenchymal stem cells involved in wound healing. Stem Cells Dev. 2017;26(13):948–63. https://doi.org/10.1089/scd.2016.0326.
Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148(3):399–408. https://doi.org/10.1016/j.cell.2012.01.021.
Semenza GL, Agani F, Booth G, Forsythe J, Iyer N, Jiang BH, et al. Structural and functional analysis of hypoxia-inducible factor 1. Kidney Intl. 1997;51(2):553–5. https://doi.org/10.1038/ki.1997.77.
Lu S, Gu X, Hoestje S, Epner DE. Identification of an additional hypoxia responsive element in the glyceraldehyde-3-phosphate dehydrogenase gene promoter. Biochimica et Biophysica Acta (BBA) Gene Struct Express. 2002;1574(2):152–6. https://doi.org/10.1016/s0167-4781(01)00359-1.
Higashimura Y, Nakajima Y, Yamaji R, Harada N, Shibasaki F, Nakano Y, et al. Up-regulation of glyceraldehyde-3-phosphate dehydrogenase gene expression by HIF-1 activity depending on Sp1 in hypoxic breast cancer cells. Arch Biochem Biophys. 2011;509(1):1–8. https://doi.org/10.1016/j.abb.2011.02.011.
Graven KK, Troxler RF, Kornfeld H, Panchenko MV, Farber HW. Regulation of endothelial cell glyceraldehyde-3-phosphate dehydrogenase expression by hypoxia. J Biol Chem. 1994;269(39):24446–53. https://doi.org/10.1016/S0021-9258(19)51104-8.
Wenger RH, Stiehl DP, Camenisch G. Integration of oxygen signaling at the consensus HRE. Science STKE. 2005;2005(306):re12. https://doi.org/10.1126/stke.3062005re12.
Dengler VL, Galbraith MD, Espinosa JM. Transcriptional regulation by hypoxia inducible factors. Crit Rev Biochem Mol Biol. 2014;49(1):1–15. https://doi.org/10.3109/10409238.2013.838205.
Yang Y, Fan W, Zhu L, Zhao T, Ma L, Wu Y, et al. Effects of hypoxia on mRNA expression of housekeeping genes in rat brain tissue and primary cultured neural cells. Front Med China. 2008;2:239–43. https://doi.org/10.1007/s11684-008-0045-7.
Bakhashab S, Lary S, Ahmed F, Schulten HJ, Bashir A, Ahmed FW, et al. Reference genes for expression studies in hypoxia and hyperglycemia models in human umbilical vein endothelial cells. G3 (Bethesda). 2014;4(11):2159–65. https://doi.org/10.1534/g3.114.013102.
Moein S, Javanmard S, Abedi M, Izadpanahi M, Gheisari Y. Identification of appropriate housekeeping genes for gene expression analysis in long-term hypoxia-treated kidney cells. Adv Biomed Res. 2017;6:15. https://doi.org/10.4103/2277-9175.200790.
Fink T, Lund P, Pilgaard L, Rasmussen JG, Duroux M, Zachar V. Instability of standard PCR reference genes in adipose-derived stem cells during propagation, differentiation and hypoxic exposure. BMC Mol Biol. 2008;9:98. https://doi.org/10.1186/1471-2199-9-98.
Foldager C, Munir S, Ulrik-Vinther M, Søballe K, Bünger C, Lind M. Validation of suitable house keeping genes for hypoxia-cultured human chondrocytes. BMC Mol Biol. 2009;10:94. https://doi.org/10.1186/1471-2199-10-94.
Baddela VS, Baufeld A, Yenuganti VR, Vanselow J, Singh D. Suitable housekeeping genes for normalization of transcript abundance analysis by real-time RT-PCR in cultured bovine granulosa cells during hypoxia and differential cell plating density. Reprod Biol Endocrinol. 2014;12:118. https://doi.org/10.1186/1477-7827-12-118.
Yao L, Chen X, Tian Y, Lu H, Zhang P, Shi Q, et al. Selection of housekeeping genes for normalization of RT-PCR in hypoxic neural stem cells of rat in vitro. Mol Biol Rep. 2012;39(1):569–76. https://doi.org/10.1007/s11033-011-0772-8.
Zhong H, Simons JW. Direct comparison of GAPDH, β-actin, cyclophilin, and 28S rRNA as internal standards for quantifying RNA levels under hypoxia. Biochem Biophys Res Commun. 1999;259(3):523–6. https://doi.org/10.1006/bbrc.1999.0815.
de Kok JB, Roelofs RW, Giesendorf BA, Pennings JL, Waas ET, Feuth T, et al. Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab Invest. 2005;85(1):154–9. https://doi.org/10.1038/labinvest.3700208.
Said HM, Hagemann C, Stojic J, Schoemig B, Vince GH, Flentje M, et al. GAPDH is not regulated in human glioblastoma under hypoxic conditions. BMC Mol Biol. 2007;8:55. https://doi.org/10.1186/1471-2199-8-55.
Bielecka ZF, Malinowska A, Brodaczewska KK, Klemba A, Kieda C, Krasowski P, et al. Hypoxic 3D in vitro culture models reveal distinct resistance processes to TKIs in renal cancer cells. Cell Biosci. 2017;7:71. https://doi.org/10.1186/s13578-017-0197-8.
Grayson WL, Zhao F, Bunnell B, Ma T. Hypoxia enhances proliferation and tissue formation of human mesenchymal stem cells. Biochem Biophys Res Commun. 2007;358(3):948–53. https://doi.org/10.1016/j.bbrc.2007.05.054.
Ohnishi S, Yasuda T, Kitamura S, Nagaya N. Effect of hypoxia on gene expression of bone marrow-derived mesenchymal stem cells and mononuclear cells. Stem Cells. 2007;25:1166–77. https://doi.org/10.1634/stemcells.2006-0347.
Isik B, Thaler R, Goksu BB, Conley SM, Al-Khafaji H, Mohan A, et al. Hypoxic preconditioning induces epigenetic changes and modifies swine mesenchymal stem cell angiogenesis and senescence in experimental atherosclerotic renal artery stenosis. Stem Cell Res Therap. 2021;12:240. https://doi.org/10.1186/s13287-021-02310-z.
Vandesompele J, de Preter K, Pattyn F, Poppe B, van Roy N, de Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034.1.
Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–50. https://doi.org/10.1158/0008-5472.can-04-0496.
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26(6):509–15. https://doi.org/10.1023/B:BILE.0000019559.84305.47.
Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. 2006;7:33. https://doi.org/10.1186/1471-2199-7-33.
Xie F, Xiao P, Chen D, Xu L, Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol. 2012;80:75–84. https://doi.org/10.1007/s11103-012-9885-2.
Tang X, Zhang N, Si H, Calderón-Urrea A. Selection and validation of reference genes for RT-qPCR analysis in potato under abiotic stress. Plant Methods. 2017;13:85. https://doi.org/10.1186/s13007-017-0238-7.
Lemma S, Avnet S, Meade M, Chano T, Baldini N. Validation of suitable housekeeping genes for the normalization of mRNA expression for studying tumor acidosis. Int J Mol Sci. 2018;19(10):2930. https://doi.org/10.3390/ijms19102930.
Wang Z, Lyu Z, Pan L, Zeng G, Randhawa P. Defining housekeeping genes suitable for RNA-seq analysis of the human allograft kidney biopsy tissue. BMC Med Genomics. 2019;12(1):86. https://doi.org/10.1186/s12920-019-0538-z.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25(4):402–8. https://doi.org/10.1006/meth.2001.1262.
Valente V, Teixeira SA, Neder L, Okamoto OK, Oba-Shinjo SM, Marie SK, et al. Selection of suitable housekeeping genes for expression analysis in glioblastoma using quantitative RT-PCR. BMC Mol Biol. 2009;10:17. https://doi.org/10.1186/1471-2199-10-17.
Savino TM, Bastos R, Jansen E, Hernandez-Verdun D. The nucleolar antigen Nop52, the human homologue of the yeast ribosomal RNA processing RRP1, is recruited at late stages of nucleologenesis. J Cell Sci. 1999;112(Pt 12):1889–900.
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–22. https://doi.org/10.1373/clinchem.2008.112797.
Solaini G, Baracca A, Lenaz G, Sgarbi G. Hypoxia and mitochondrial oxidative metabolism. Biochim Biophys Acta. 2010;1797(6–7):1171–7. https://doi.org/10.1016/j.bbabio.2010.02.011.
Lok CN, Ponka P. Identification of a hypoxia response element in the transferrin receptor gene. J Biol Chem. 1999;274(34):24147–52. https://doi.org/10.1074/jbc.274.34.24147.
Redmond SL, Levin B, Heel KA, Atlas MD, Marano RJ. Phenotypic and genotypic profile of human tympanic membrane derived cultured cells. J Mol Histol. 2011;42(1):15–25. https://doi.org/10.1007/s10735-010-9303-5.
Andrews S, Krueger F, Segonds-Pichon A, Biggins L, Krueger C, Wingett S. FastQC: A quality control tool for high throughput sequence data. Babraham Institute. 2012. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 11 July 2019.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. https://doi.org/10.1093/bioinformatics/btu170.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–60. https://doi.org/10.1038/nmeth.3317.
Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31(12):2032–4. https://doi.org/10.1093/bioinformatics/btv098.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30. https://doi.org/10.1093/bioinformatics/btt656.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. https://doi.org/10.1186/s13059-014-0550-8.
Zhu A, Ibrahim JG, Love MI. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics. 2019;35(12):2084–92. https://doi.org/10.1093/bioinformatics/bty895.
Acknowledgements
The authors would like to acknowledge Australian Genome Research Facility for performing the sequencing.
Funding
This work was supported by the Ear Science Institute Australia and the Gift of Hearing Appeal. HTO was supported by an Australian Government Research Training Program (RTP) Scholarship, and the Sarich and Stokes Research Scholarship.
Author information
Authors and Affiliations
Contributions
The project was conceptualised by HTO and RJD. HTO and RJD contributed to the design of the methodology. CMP contributed to the experiment design for PCR validation. HTO performed all experiments and data analyses, and was a major contributor in writing the manuscript. CMP and RJD substantially revised the manuscript. All authors contributed to the interpretation of data, read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not Applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
List of 114 hypoxia-inducible genes, gene symbol, and gene ID.
Additional file 2.
Preliminary qRT-PCR data analysis of RPL13A, TBP, GAPDH, B2M, VEGFA, using RNA extracted of hADSC cultured in normal and hypoxic conditions.
Additional file 3.
Ranking of normalised counts of RNAseq data of Nx-hADSC and Hx-hADSC curated for selected HKG candidates and calculated for CV.
Additional file 4.
R markdown script of differential expression analysis of RNAseq data using DESeq2.
Additional file 5.
Summary of hADSC donor information and cell characterisation used in this study.
Additional file 6.
Quality control and qualification results of RNA extracted from hADSC.
Additional file 7.
RNAseq processing efficiencies represented as percentage counts of original sequenced reads
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ong, H.T., Prêle, C.M. & Dilley, R.J. Using RNA-seq to identify suitable housekeeping genes for hypoxia studies in human adipose-derived stem cells. BMC Mol and Cell Biol 24, 16 (2023). https://doi.org/10.1186/s12860-023-00475-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12860-023-00475-4