Abstract
Background
Transcriptomic profiling of ex vivo cultured human limbal epithelial stem cells (hLESCs) will foster better understanding of corneal physiology and novel treatment paradigms to limbal stem cell deficiency (LSCD). However, currently such profiling studies are hampered due to difficulties with producing sufficient amounts of intact mRNA for deep RNA sequencing (RNA-seq) from subpopulations sorted on the basis of co-expression of membrane and intracellular antigens by fluorescence-activated cell sorting (FACS).
Methods
To address this problem, we systematically analyzed the critical steps, and found that ethanol fixation together with optimized downstream procedures provided a pipeline that yielded high quality total RNA in amounts to readily support the RNA-seq procedure, while still preserving good discrimination between the individual hLESC immunophenotypes.
Results
The average RNA integrity number (RIN) was 7.7 ± 0.4, and the average yield was 4.6 ± 1.7 pg of RNA per cell. The sequencing analysis of the isolated RNA produced high quality data with more than 70% of read pairs mapping uniformly to the reference genome and 80% of bases with a Phred score of at least 30.
Conclusion
In this study, we developed a reliable FACS-based procedure using ethanol as a fixative that would support accurate isolation of limbal epithelial progenitor subpopulations along with RNA yield and quality sufficient to enable deep transcriptomic profiling.
Similar content being viewed by others
Background
Corneal epithelial disease due to limbal stem cell deficiency (LSCD) is a major cause of blindness worldwide [1]. The use of ex vivo expanded human limbal epithelial stem cells (hLESCs) for transplantation [2] has been a remarkable success, however with a failure rate of 30% there is still room for improvement [3]. Previously, advanced techniques, including laser capture microdissection and deep RNA sequencing (RNA-seq), provided an unprecedented level of detail of molecular networks involved in the regulation of different limbal niche compartments in vivo [4]. Similar level of information regarding the cultured hLESCs is needed to understand how the ex vivo phenotype departs from the in vivo counterpart, and, importantly, to devise procedures whereby hLESCs can be expanded while preserving the properties they had in situ. To this end, a detailed transcriptomic analysis of specific hLESC lineages is warranted.
To achieve this goal, defined hLESC subpopulations need to be identified and isolated, and ultimately yield a high quality RNA. In the face of difficulties to identify an archetypal stem cell profile, the stemness in the cultured hLESCs is currently believed to be associated with the surface ATP-binding cassette sub-family B member 5 (ABCB5) [5] and/or intranuclear transformation-related protein 63 (p63) [6] antigens, in the absence of the cytoplasmic differentiation marker cytokeratin 3 (CK3) [7]. Consequently, any sorting attempt is contingent on a complex immunostaining procedure for both extra- and intracellularly located molecules that must yield accurate and reliable discrimination of different epitopes, and simultaneously preserve the RNA.
The fixation and permeabilization steps have in particular been shown to have an adverse effect on the RNA integrity [8,9,10], but the sorting procedure also presents a formidable obstacle due to challenges with securing an RNase-free environment and minimizing the physical stress imposed on the cells [11,12,13].
There is only scanty literature dealing with transcriptomic profiling of cells sorted by FACS on the basis of simultaneous surface and intracellular immunostaining. Incidentally, most of these reports are based on the use of formaldehyde as a fixative [14,15,16,17] but also methanol [18]. Our own results unfortunately could not lend credence to the usefulness of these fixatives in similar scenarios due to a high level of RNA degradation (unpublished data). Along these lines, evidence has previously been produced to show that formaldehyde results in a decrease of RNA integrity when used for transcriptional analysis of histological specimens [8,9,10]. The formation of irreversible cross-links between nucleic acids and proteins is believed to play a major role [8]. To avoid cross-linking, ethanol has been assayed, and turned out to compare satisfactory in terms of RNA yield and integrity [19,20,21]. Ethanol thus appears as a good candidate in a complex setup, where high quality RNA needs to be obtained in the face of multistep procedure involving fixation, permeabilization, labeled antibodies, and FACS sorting. Since there is no evidence that such trial would be carried out previously, we attempted to invoke ethanol in conjunction with all other specific steps to outline a novel pipeline to support RNA-seq and applied it to cultured hLESCs.
Methods
Cell Culture
The hLESCs isolation and culture protocol was optimized based on our previous report [22]. Human corneal scleral ring remnants (donors aged 22–86 years, 64% were men, absence of corneal disease) after keratoplasty procedures were obtained from the Department of Ophthalmology, Aarhus University Hospital (Arhus, Denmark), and handled according to Danish legislature. In brief, the rings from 10 to 12 randomly chosen donors were first rinsed with sterile phosphate buffered saline (sPBS, Gibco™, Life Technologies, Naerum, Denmark) supplemented with 100 units/ml penicillin and 100 μg/ml streptomycin (Life Technologies). After scraping off the endothelial side, the rings were incubated with 2.4 units/ml dispase II (Life Technologies) in sPBS at 37 °C for 1 h. Cells were then removed from the epithelial side, pooled briefly and pelleted at 300 g for 5 min, incubated with TrypLE Express (Life Technologies) at 37 °C for 8 min, and finally purified through a 70 μm cell-strainer (BD Biosciences, San Jose, CA). The resulting cell suspension was seeded directly into a T25 culture flask (Corning CellBIND; Sigma–Aldrich, Copenhagen, Denmark) at a density of 1000 to 1500 cells/cm2 in complete Keratinocyte-SFM (Life Technologies). The medium was changed every 2–3 days. At 80–90% confluency, the cells were trypsinized using TrypLE Express at 37 °C for 10 min, filtered through strainer, washed and resuspended in sPBS. Single cell suspensions of 0.5 to 1.5 × 106 cells were aliquoted into 2.0 ml RNase-free microfuge tubes (Ambion, Naerum, Denmark) for subsequent immunofluorescence staining. The isolation and expansion process were repeated at 3 distinct time points.
Fixation and Permeabilization
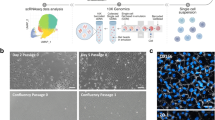
For optimization, two fixation and permeabilization methods were compared. The first method included fixation with 4% formaldehyde (Applichem, Esbjerg, Denmark) at room temperature for 15 min and subsequent permeabilization with 0.1% saponin (Sigma–Aldrich) in PBS supplemented with 1:100 Rnasin Plus Rnase inhibitor (Promega, Roskilde, Denmark) at 4 °C for 30 min. The second method was a one-step fixation and permeabilization using 70% ethanol in PBS (Ethanol absolute Electran® Molecular biology grade, VWR, Søborg, Denmark) at 4 °C for 10 min (Fig. 1a).
Effect of immunostaining and FACS on the integrity of RNA. a Flow chart of the staining and sorting procedures indicating the critical steps during the FACS-mediated isolation of the subpopulations SP1–4 and the steps at which RNA integrity was analyzed. Flash-freeze indicates samples flash frozen in liquid nitrogen and stored at −80 °C after the sorting. b Qualitative integrity of total RNA from the discrete steps during the immunostaining and sorting procedure. The quantitative data are presented as mean ± SD (n = 2–4). * denotes a significant (p < 0.05) lower RIN when compared to RIN directly after fixation and permeabilization with 70% ethanol, † denotes a significant lower RIN by pairwise comparison. ABCB5 = ATP-binding cassette sub-family B member 5; CH2O = formaldehyde; C2H5OH = ethanol; CK3 = cytokeratin 3; p63 = transformation-related protein 63; LN2 = liquid nitrogen; RIN = RNA integrity number; FACS = fluorescence-activated cell sorting
Immunofluorescence Staining
The cells were first stained in suspension with direct conjugated antibody against the surface marker ABCB5 (for more information on antibodies, see Additional file 1), followed by washing with sPBS includes 50% Accumax (Sigma-Aldrich), and 25 mM HEPES (Life Technologies) at 300 g at 4 °C for 3 min and fixation and permeabilization with 70% ethanol at 4 °C for 10 min. After further washing, directly conjugated antibodies against intracellular p63 and CK3 were applied, and the staining procedure was concluded by two additional washing steps. As negative controls, non-specific isotype matching antibodies were used for p63 and CK3, and fluorescence minus one (FMO) for ABCB5. The antibodies were used in optimal dilution in PBS with 0.1% BSA (Sigma-Aldrich) and 50% Accumax, and 25 mM HEPES. After fixation and permeabilization, the staining and washing buffers were supplemented with Rnasin Plus Rnase Inhibitor diluted 1:10 and 1:100, respectively. All staining steps were performed in the dark at 4 °C for 30 min, and the volumes were kept at 100 μl for 106 cells. In order to discriminate dead cells, the LIVE/DEAD Fixable Dead Cell Stain Kit (Life Technologies) was used according to the manufacturer’s instructions. Following staining, the cells were resuspended in a sort/collect buffer consisting of the washing buffer with 10-fold diluted RNasin Plus Rnase Inhibitor in a concentration to yield from 0.5 to 2 × 106 cells per ml. The cell suspensions were then filtered through the Pre-Separation Filters (Miltenyi Biotec, Bergisch Gladbach, Germany) into 5 ml round bottom polystyrene tubes (BD Falcon, Albertslund, Denmark). All samples were left on ice in the dark until FACS.
FACS
The flow cytometric analysis and sorting were performed using a MoFlo Astrios cell sorter (Beckman Coulter, Brea, CA). In order to avoid RNase contamination, the instrument was thoroughly decontaminated with a cleaning agent (COULTER CLENZ; Beckman Coulter), RNaze ZAP (Sigma-Aldrich), 70% ethanol, and milli-Q water. After decontamination, RNaseAlert Lab Test kit (Ambion) was used to ensure the complete absence of RNase.
For phenotype analysis and gate setting, data on a minimum of 10,000 events were collected. For sorting, the Summit Software v4.3 (Beckman Coulter) was used, with the sort mode set as “purify”, and the gates set with reference to the isotype and FMO controls (Fig. 2a). To maximize the sample integrity, the sorting procedure was run at low pressure (20 Psi), a 100 μm sorting nozzle was used and the temperature of the sorting chamber as well as the flow line was maintained at 4 °C. A typical sorting run took 2 to 3 h to complete. Four subpopulations were sorted (SP1–4), consisting of cells with the following profiles ABCB5+p63+CK3+ (SP1), ABCB5+p63+CK3− (SP2), ABCB5+p63−CK3− (SP3), and ABCB5−p63+CK3− (SP4). The subpopulations were sorted into polypropylene round bottom FACS tubes (BD Falcon) pre-filled with the sort/collect buffer. After taking an aliquot for quality control, the sorted subpopulations were pelleted, the supernatant was drained completely, and the samples were flash-frozen in liquid nitrogen. Frozen cell pellets were kept at −80 °C until RNA analysis.
Effect of optimized procedure on the separation of subpopulations and the sequencing analysis of RNA quality. a Discrimination from background using top 2.5 percentile of control intensity as a cut-off is shown in representative traces for both membrane (ABCB5) and intracellular (p63 and CK3) antigens. Robust and specific sorting produced four subpopulations indicated in boldface. b The quality of reads was assessed by sequence quality (PHRED score), mapping coverage, and GC-content distribution
RNA Analysis
For quality control the integrity of the RNA was assayed at several steps during the staining and sorting procedure with the aid of the Aurum Total RNA kit (Bio-Rad, Copenhagen, Denmark) and the Agilent 2100 Bioanalyzer (Agilent Technologies, Glostrup, Denmark) both as prescribed by the manufacture (Fig. 1a).
After flash-freezing the transcriptomic analysis of the frozen subpopulations (SP1–4) was performed by AROS Applied Biotechnology (Arhus, Denmark) using the SMART-Seq v4 Ultra Low Input RNA Kit for cDNA and Low Input Library Prep Kit V2 for sequencing library preparation (both by Takara Bio, Otsu, Japan), and Illumina HiSeq 2000 instrument (Illumina Inc., San Diego, CA) for sequencing. The sequencing data were imported as paired-ends into CLC Genomic Workbench v9.0.1 (CLC bio, Aarhus, Denmark) to enable RNA-seq reports, including reads average PHRED score and mapping statistics against the annotated Homo sapiens UCSC hg19 reference genome (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/bigZips/). The alignment quality was further assessed by the Qualimap v2.2 (http://qualimap.bioinfo. cipf.es/) producing the read coverage and GC content profiles [23].
Results
Effect of Fixation and Subsequent Cell Processing on RNA
The optimization of the procedure for RNA-seq of the ex vivo expanded and FACS sorted hLESCs was done according to the paradigm illustrated in Fig. 1a. To evaluate the effect of fixation and downstream processing on the RNA quality, the samples were analyzed at each step of the protocol (Fig. 1b). No obvious effect on the visualization of ABCB5, p63, and CK3 markers in limbal epithelial cell culture was observed when treated with either 70% ethanol or 4% formaldehyde and followed by saponin as permeabilization agent (data not shown). As indicated by the electrophoretic profiles and the quantitative assessment, the fixation with 4% formaldehyde and permeabilization with 0.1% saponin resulted in practically complete disintegration of the RNA. On the other hand, implementation of 70% ethanol as a fixative, produced practically intact RNA with RIN 9.9 ± 0.1 (max RIN = 10). During the following downstream step involving the immunostaining, the RNA integrity became only marginally affected (RIN 9.4 ± 0.3), whereas the FACS sorting had a more pronounced effect, resulting in RIN 8.4 ± 0.1. Importantly from the practical point of view, we found that freezing of the sorted cells for later RNA extraction resulted in only marginally lowered RIN (7.7 ± 0.4), thus meeting the RIN recommendation for the advanced RNA sequencing platform (RIN >7).
When comparing different subpopulations, a considerable variability in RNA yield per cell was found, ranging from 2.25 to 6.37 pg of RNA per cell, with an average of 4.59 ± 1.71 from all four subpopulations. Additionally, a positive correlation between the number of lysed cells and the integrity of produced RNA was observed (Table 1). Consequently, it was important that approximately 2 × 107 cells were processed for each run so that at least 400 ng of total RNA would be harvested, which is the amount necessary to guarantee a high quality sequencing data.
FACS Sorting of Limbal Epithelial Progenitors and Sequencing Analysis
The optimized immunostaining procedure enabled robust and specific identification of the surface and intracellular epitopes as indicated by overlays with control histograms (Fig. 2a). Based on co-expression of individual markers, four distinct subpopulations (SP1–4) could then be sorted out. As shown in the Venn diagram, the cells expressing only a single stemness-associated marker ABCB5 or p63, or combination thereof, comprised only 13.3% of the hLESC culture. The differentiation marker CK3 was expressed alone or in combination with stemness markers in 44.3% of the cells. Repertoires highlighted in bold were sorted for subsequent RNA-seq.
The performance of the sequencing was assessed by assessing the overall read quality, the mapping statistics and read coverage, and the GC content (Fig. 2b, Table 2). The average read count was 114.03 × 106, which corresponds well with the expected sequencing sensitivity for genes expressed at low levels. The PHRED quality score (>80%) was higher than 30 for 80% of all base calls. For mapped reads, the average GC content was 49.25% and the GC content per sample displayed a normal distribution. After alignment, on average, 82.67% of reads were mapped in pairs to the reference genome (human genome hg19), and 86.96% of total mapped reads were mapped to exonic regions. Reads coverage profile over gene body was obtained by scaling all transcripts to 100 nt and calculating the number of reads covering each nucleotide position. The result showed uniform gene body coverage. Raw sequencing data were deposited in the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.Nih.gov/geo/) under the accession number PRJNA387095 and ID 387095.
Statistics
The data are presented as mean ± SD from three independent experiments entailing a total of 2–4 biological replicates. One-way ANOVA (LSD) of the SPSS 24.0 package (SPSS, Chicago, IL) was used to test for differences between the samples, and P < 0.05 was set to discriminate statistical significance.
Discussion
During the last decade, there has been an impressive advancement in the tools for transcriptome analysis in the scientific community and in our laboratory, from the rather simple membrane arrays allowing simultaneous analysis of a couple of hundreds of different genes [24], to more advanced microarrays enabling analysis of several hundreds of thousand genes [25]. In addition to these hybridization arrays, that are based on genes known a priori and that require a somewhat high amount of input RNA, the more advanced techniques such as deep RNA sequencing (RNA-seq), has facilitated a global analysis of the transcriptome permitting detection of alternative transcripts, with a high dynamic range, and from a smaller amount of RNA [26]. Notwithstanding, as with all techniques, the quality of the input RNA is imperative for reliable results [27].
For the sorting of heterogeneous cell populations, we rely on both extracellular and intracellular markers, and therefore need to fix and permeabilize the cells prior to staining and sorting. Formaldehyde has traditionally been used as a fixative in histology and immunochemistry due to its ability to preserve cell morphology. This is due to the formation of protein-protein and protein-nucleic acid cross-links involving methylene bridges (-CH2-) [28]. However, the use of formaldehyde as a fixative in molecular biology has been questioned, since it has been suspected to cause RNA degradation and/or making it difficult to extract RNA due to cross-linking with proteins or chemical modifications [29]. In recent years, multiple studies have indicated that non-cross-linking alcohol-based fixatives, such as ethanol, allow for the isolation of RNA with better integrity [19,20,21]. This is due to the fact that ethanol does not support cross-linking but allows for minor chemical modifications to the nucleic acids [8]. Our study also corroborated the superiority of ethanol, since no tangible RNA degradation was seen after the fixation and permeabilization, and during the following downstream steps, the RNA quality was preserved to support high quality sequence data.
Another major challenge in this study was to protect the RNA against the RNase-mediated degradation. The cells in our protocol were permeabilized to reveal intracellular antigens, which allowed for an increased exposure to contaminating extracellular RNase. RNase inhibitor was therefore included in all solutions following fixation and permeabilization, and rigorous cleaning was implemented to decontaminate all surfaces coming into contact with the cells. As another measure to reduce the exposure to RNase, the buffering capacity of the solutions was boosted by adding a non-phosphate HEPES system and a gentle cell detachment solution (Accumax) was implemented with the aim to minimize the cell death. All additives used in this study were previously shown to have no adverse effect on RNA integrity [11]. Since cells undergoing FACS can experience physical stress [12], RNA integrity can be further improved by decreasing the fluid pressure and/or increasing nozzle diameter [13], and keeping the cells at a lowered temperature. It is also recommended that the cells are directly sorted into RNA extraction buffer [30], however, when dealing with a large number of cells, the high volume of sheet fluid can result in undesirable dilution of RNA extraction buffer and possibly reduce RNA recovery. Thus in our study, the cells were deposited directly in the sPBS-based buffer, pelleted and frozen for storage.
Since degration of RNA will greatly affect the coverage of transcripts, it is of great importance to control quality of input material before transcriptomic profiling by assessing raw reads quality and mapping to the reference genome [31]. Before aligment, the quality of sample reads was assessed by length, GC content, quality score and duplicate sequences. After alignment, read mapping and read coverage was checked to further ensure the quality of the sequencing data [32]. Despite ongoing effort to improve the RNA sequencing quality control, it remains difficult to quantify the effect of RNA degradation on the sequencing performance. Transcript integrity number (TIN) was proposed to measure RNA degration at the transcript level [33]. Nevertheless, this measure was derived from tissue studies and further investigation is necessary to confirm its value in the cell culture scenario.
Conclusions
Purification and characterization of progenitor cells in human corneal epithelium remains a challenge since the stem cells constitute only a small fraction (< 10%) of the cultured limbal epithelial cell population [34] and their exact immunophenotypical profile remains elusive. Thus a panel of proposed putative stem/progenitor markers are often applied to detect hLESCs. From the perspective point of view, our novel approach demonstrates the possibility of sorting corneal phenotypes based on p63, ABCB5, and CK3 for RNA-seq, which holds promise to reveal whether these markers are associated with gene activation that is characteristic of limbal stem or precursor cells. Further investigations are undoubtedly warranted to confirm utility of this approach beyond the cultured hLESCs.
Abbreviations
- ABCB5:
-
ATP-binding cassette sub-family B member 5
- CK3:
-
Cytokeratin 3
- FACS:
-
Fluorescence-activated cell sorting
- FMO:
-
Fluorescence minus one
- hLESCs:
-
Human limbal epithelial stem cells
- LSCD:
-
Limbal stem cell deficiency
- p63:
-
Transformation-related protein 63
- RIN:
-
RNA integrity number
- RNA:
-
Ribonucleic acid
- RNA-seq:
-
RNA sequencing
- SD:
-
Standard deviation
- SP:
-
Subpopulation
- TIN:
-
Transcript integrity number
References
Liang L, Sheha H, Li J, Tseng SCG. Limbal stem cell transplantation: new progresses and challenges. Eye. 2009;23:1946–53.
Pellegrini G, Traverso CE, Franzi AT, Zingirian M, Cancedda R, de Luca M. Long-term restoration of damaged corneal surfaces with autologous cultivated corneal epithelium. Lancet. 1997;349:990–3.
Haagdorens M, Van Acker SI, Van Gerwen V, Ní Dhubhghaill S, Koppen C, Tassignon M-J, et al. Limbal stem cell deficiency: current treatment options and emerging therapies. Stem Cells Int. 2016;2016:1–22.
Bath C, Muttuvelu D, Emmersen J, Vorum H, Hjortdal J, Zachar V, et al. Transcriptional dissection of human Limbal niche compartments by massive parallel sequencing. PLoS One. 2013;8:e64244.
Ksander BR, Kolovou PE, Wilson BJ, Saab KR, Guo Q, Ma J, et al. ABCB5 is a limbal stem cell gene required for corneal development and repair. Nature. 2014;511:353–7.
Pellegrini G, Dellambra E, Golisano O, Martinelli E, Fantozzi I, Bondanza S, et al. P63 identifies keratinocyte stem cells. Proc Natl Acad Sci U S A. 2001;98:3156–61.
Kiritoshi A, SundarRaj N, Thoft RA. Differentiation in cultured limbal epithelium as defined by keratin expression. Investig Ophthalmol Vis Sci. 1991;32:3073–7.
Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161:1961–71.
Cox ML, Schray CL, Luster CN, Stewart ZS, Korytko PJ, Khan M. KN, et al. assessment of fixatives, fixation, and tissue processing on morphology and RNA integrity. Exp Mol Pathol. 2006;80:183–91.
Cox ML, Eddy SM, Stewart ZS, Kennel MR, Man MZ, Paulauskis JD, et al. Investigating fixative-induced changes in RNA quality and utility by microarray analysis. Exp Mol Pathol. 2008;84:156–72.
Diez C, Bertsch G, Simm A. Isolation of full-size mRNA from cells sorted by flow cytometry. J Biochem Biophys Methods. 1999;40:69–80.
Mollet M, Godoy-Silva R, Berdugo C, Chalmers JJ. Acute hydrodynamic forces and apoptosis: a complex question. Biotechnol Bioeng. 2007;98:772–88.
Suh TK, Schenk JL, Seidel GE. High pressure flow cytometric sorting damages sperm. Theriogenology. 2005;64:1035–48.
Hrvatin S, Deng F, O’Donnell CW, Gifford DK, Melton DAMARIS. Method for analyzing RNA following intracellular sorting. PLoS One. 2014;9:e89459.
Nilsson H, Krawczyk KM, Johansson ME. High salt buffer improves integrity of RNA after fluorescence-activated cell sorting of intracellular labeled cells. J Biotechnol. 2014;192(Part A):62–5.
Russell JN, Clements JE, Gama L. Quantitation of gene expression in formaldehyde-fixed and fluorescence-activated sorted cells. PLoS One. 2013;8:e73849.
Jeyapalan JC, Sedivy JM. How to measure RNA expression in rare senescent cells expressing any specific protein such as p16Ink4a. Aging (Albany NY). 2013;5:120–9.
Yamada H, Maruo R, Watanabe M, Hidaka Y, Iwatani Y, Takano T, Messenger RNA. Quantification after fluorescence activated cell sorting using intracellular antigens. Biochem Biophys Res Commun. 2010;397:425–8.
Groelz D, Sobin L, Branton P, Compton C, Wyrich R, Rainen L. Non-formalin fixative versus formalin-fixed tissue: a comparison of histology and RNA quality. Exp Mol Pathol. 2013;94:188–94.
JMF S, Perlaky L, Li X-N, HCE L, Antalffy B, Armstrong D, et al. Comparison of ethanol versus formalin fixation on preservation of histology and RNA in laser capture microdissected brain tissues. Brain Pathol. 2004;14:175–82.
Milcheva R, Janega P, Celec P, Russev R, Babál P. Alcohol based fixatives provide excellent tissue morphology, protein immunoreactivity and RNA integrity in paraffin embedded tissue specimens. Acta Histochem. 2013;115:279–89.
Bath C, Yang S, Muttuvelu D, Fink T, Emmersen J, Vorum H, et al. Hypoxia is a key regulator of limbal epithelial stem cell growth and differentiation. Stem Cell Res. 2013;10:349–60.
Okonechnikov K, Conesa A, García-Alcalde F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2015;32:292–4.
Fink T, Ebbesen P, Zachar V. Quantitative gene expression profiles of human liver-derived cell lines exposed to moderate hypoxia. Cell Physiol Biochem. 2001;11:105–14.
Lund P, Pilgaard L, Duroux M, Fink T, Zachar V. Effect of growth media and serum replacements on the proliferation and differentiation of adipose-derived stem cells. Cytotherapy. 2009;11:189–97.
Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63.
Bath C, Fink T, Vorum H, Hjortdal J, Zachar V. Technical brief: optimized pipeline for isolation of high-quality RNA from corneal cell subpopulations. Mol Vis 2014;20 April 2013:797–803.
Thavarajah R, Mudimbaimannar VK, Elizabeth J, Rao UK, Ranganathan K. Chemical and physical basics of routine formaldehyde fixation. J Oral Maxillofac Pathol. 2012;16:400–5.
Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999;27:4436–43.
Tighe S, Held MA. Isolation of total rna from transgenic mouse melanoma subsets using fluorescence-activated cell sorting. Methods Mol Biol. 2010;632:27–44.
Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13.
Li X, Nair A, Wang S, Wang L. Quality control of RNA-seq experiments. In: RNA. Bioinformatics. 2015:137–46.
Wang L, Nie J, Sicotte H, Li Y, Eckel-Passow JE, Dasari S, et al. Measure transcript integrity using RNA-seq data. BMC Bioinformatics. 2016;17:58.
Lavker RM, Dong G, Cheng SZ, Kudoh K, Cotsarelis G, Sun TT. Relative proliferative rates of limbal and corneal epithelia. Implications of corneal epithelial migration, circadian rhythm, and suprabasally located DNA-synthesizing keratinocytes. Investig Ophthalmol Vis Sci. 1991;32:1864–75.
Acknowledgments
We thank Esben Nielsen and Henrik Sejersen at the Department of Ophthalmology, Aarhus University Hospital for the collection of experimental materials.
Funding
Publication of this article was funded by Aalborg University.
Availability of Data and Materials
The datasets generated and/or analysed during the current study are available in the Gene Expression Omnibus repository, [http://www.ncbi.nlm.Nih.gov/geo/] under the accession number PRJNA387095 and ID 387095.
Author information
Authors and Affiliations
Contributions
LL: Study conception and design, acquisition of data, analysis and interpretation of data. FMN: Acquisition and analysis of data. SER: Analysis and interpretation of data, critical revision. JE: Analysis and interpretation of data. TF: Drafting of manuscript, critical revision. JH: Study conception and design, critical revision. CB: Study conception and design, critical revision. VZ: Study conception and design, analysis and interpretation of data, drafting of manuscript, critical revision. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Human corneal scleral ring remnants, otherwise meant for destruction after keratoplasty procedure, served as expirimental materials in this study and were handled according to Danish legislature after guidance from the local ethical committee and the Danish Health and Medicines Agency.
Consent for Publication
Not applicable.
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
List of antibodies. (DOCX 12 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Liu, L., Nielsen, F.M., Riis, S.E. et al. Maintaining RNA Integrity for Transcriptomic Profiling of Ex Vivo Cultured Limbal Epithelial Stem Cells after Fluorescence-Activated Cell Sorting (FACS). Biol Proced Online 19, 15 (2017). https://doi.org/10.1186/s12575-017-0065-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12575-017-0065-2