Abstract
Sakuranetin plays a key role as a phytoalexin in plant resistance to biotic and abiotic stresses, and possesses diverse health-promoting benefits. However, mature rice seeds do not contain detectable levels of sakuranetin. In the present study, a transgenic rice plant was developed in which the promoter of an endosperm-specific glutelin gene OsGluD-1 drives the expression of a specific enzyme naringenin 7-O-methyltransferase (NOMT) for sakuranetin biosynthesis. The presence of naringenin, which serves as the biosynthetic precursor of sakuranetin made this modification feasible in theory. Liquid chromatography tandem mass spectrometry (LC–MS/MS) validated that the seeds of transgenic rice accumulated remarkable sakuranetin at the mature stage, and higher at the filling stage. In addition, the panicle blast resistance of transgenic rice was significantly higher than that of the wild type. Specially, the matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) imaging was performed to detect the content and spatial distribution of sakuranetin and other nutritional metabolites in transgenic rice seeds. Notably, this genetic modification also did not change the nutritional and quality indicators such as soluble sugars, total amino acids, total flavonoids, amylose, total protein, and free amino acid content in rice. Meanwhile, the phenotypes of the transgenic plant during the whole growth and developmental periods and agricultural traits such as grain width, grain length, and 1000-grain weight exhibited no significant differences from the wild type. Collectively, the study provides a conceptual advance on cultivating sakuranetin-rich biofortified rice by metabolic engineering. This new breeding idea may not only enhance the disease resistance of cereal crop seeds but also improve the nutritional value of grains for human health benefits.
Similar content being viewed by others
Introduction
Sakuranetin (4', 5-dihydroxy-7-methoxyflavanone) is an inducible secondary metabolite, that belongs to the class of flavonoids known as flavanones. It was first extracted from the bark of Chinese cherries (Prunus pseudocerasus), and subsequently identified in a series of rice studies as a new phytoalexin with brilliant activity against rice blast (Kodama et al. 1992; Hasegawa et al. 2014; Murata et al. 2020). In addition to the accumulation in rice in response to Magnaporthe oryzae infection, sakuranetin also possesses broad-spectrum resistance to a wide range of phytopathogenic fungi (e.g., Rhizoctonia solani and Bipolaris oryzae) and bacteria (e.g., Xanthomonas oryzae pv. oryzae, Xanthomonas oryzae pv. oryzicola, and Burkholderia glumae) (Hasegawa et al. 2014; Park et al. 2014). Moreover, UV radiation, CuCl2, jasmonic acid, brassinosteroid, and brown planthopper (BPH) attack also induced the accumulation of sakuranetin in rice (Shimizu et al. 2012b; Ogawa et al. 2017; Liu et al. 2023; He et al. 2024). In fact, beyond acting as a phytoalexin in plants, sakuranetin holds tremendous promise as a nutraceutical or pharmaceutical agent, as well as an active ingredient in skincare products, due to its exceptional antiviral, anticancer, anti-inflammatory, antiparasitic, antioxidant, and anti-allergic properties (Melo et al. 2018; Stompor 2020; Junaid et al. 2022). For example, sakuranetin is effective against rhinoviruses, influenza B, B16BL6 melanoma, ESCC (esophageal squamous cell carcinoma), Colo 320 (colon cancer), leishmaniasis, and Chagas disease (Ugocsai et al. 2005; Hong and Ying 2015; Drira and Sakamoto 2016; Choi 2017; Kwon et al. 2018; Santana et al. 2019). The biosynthetic pathway of sakuranetin in rice has been clearly studied, as illustrated in Fig. 1, which is synthesized through the flavonoid branch of the general phenylpropanoid pathway. Enzymes commonly found in flavonoid metabolic pathways, including phenylalanine ammonialyase (PAL), cinnamic acid 4-hydrolase (C4H), coumarin-CoA ligase (4CL), chalcone synthase (CHS), and chalcone isomerase (CHI), are involved in the synthesis of sakuranetin in rice. Furthermore, the methyltransferase NOMT (Naringenin 7-O-methyltransferase) plays a crucial role as a specific key enzyme that catalyzes the biosynthesis of sakuranetin from the naringenin (Shimizu et al. 2012a; Rakwal et al. 2000). Even though the pathogen resistance function of sakuranin in rice leaves has been widely concerned, previous metabolomics found that sakuranetin was undetectable in mature rice seeds (Shi et al. 2022; Tang et al. 2022).
The pathways of sakuranetin biosynthesis in rice. Metabolites are indicated in black and different enzymes are indicated in color. PAL, phenylalanine ammonialyase; C4H, cinnamic acid 4-hydrolase; 4CL, coumarin-CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; FNS, flavone synthase; F3'H, flavonoid-3'-hydroxylase; F3H, flavanone-3-hydroxylase; NOMT, Naringenin 7-O-methyltransferase
In recent years, many studies have used metabolic engineering and synthetic biology to modify flavonoid metabolic and regulatory pathways to generate biofortified crops. The overexpression of MYB transcription factors has been shown to enhance anthocyanin content significantly, resulting in the production of pigment-rich crops such as purple-fleshed sweet potato, purple cauliflower, red-fleshed apple, blood orange, and purple pummelo (Mano et al. 2007; Espley et al. 2009; Chiu et al. 2010; Butelli et al. 2012; Huang et al. 2018). Besides, driving a functional SlAN2-like gene through fruit-specific promoters also leads to the activation of the entire anthocyanin biosynthesis pathway in tomatoes and massive accumulation of anthocyanins in both the peel and flesh (Sun et al. 2020). In addition to being widely used in tissues such as fruits, leaves, and flowers of fruits and vegetables, metabolic engineering to modify flavonoid metabolic pathways has also been applied to rice endosperm. Flavonoids are absent or very low in the rice endosperm, although anthocyanins are abundant in the pericarp of natural black and red rice grains. Ogo et al. successfully produced a transgenic rice plant capable of synthesizing several classes of flavonoids in its seeds, which was accomplished by introducing multiple genes encoding enzymes involved in flavonoid synthesis, from phenylalanine to the target flavonoids (Ogo et al. 2013). Similarly, Zhu et al. produced “Purple Endosperm Rice” (called Zijingmi in Chinese) by accumulating anthocyanins in endosperm using a high-efficiency multigene vector system TGSII, and this binary vector was constructed containing two regulatory genes and six structural genes for endosperm-specific anthocyanin synthesis (Zhu et al. 2017).
As we all know, rice seeds not only provide energy for plant growth and development, but are also an essential source of various nutrients for humans. Remarkably, rice endosperm has emerged as an ideal bioreactor for plant molecular farming to produce and store the recombinant proteins (antigens and vaccines/edible vaccines, pharmaceutical proteins and peptides, and antibodies) and bioactive metabolites (vitamins and minerals, carotenoids, and flavonoids). This is primarily due to its advantages, including rich genetic and bioinformatic resources, strong expression of complex proteins, ease of extraction and processing, low production costs, and few biosafety risks (Zhu et al. 2022). For instance, the expression of daffodil or maize phytoene synthase gene PSY in the rice endosperm can produce different amounts of beta-carotene that are lacking in rice seeds, thus cultivating the famous “Golden Rice” rich in provitamin A (Ye et al. 2000; Paine et al. 2005). Zhu et al. used TGSII system led to de novo astaxanthin biosynthesis in rice endosperm by co-express PSY, CrtI, BHY, and BKT gene, producing astaxanthin-enriched “Astaxanthin Rice” (called Chijingmi in Chinese) (Zhu et al. 2018). Using the 2A-mediated polycistronic method, Ha et al. achieved the biosynthesis of zeaxanthin, astaxanthin, and capsanthin in rice endosperm by stepwise pathway engineering (Ha et al. 2019). However, the above modifications to rice endosperm are all from the nutrition and health perspective, without considering plant resistance. Sakuranetin, the sole flavonoid phytoalexin with favorable implications for human health, has the potential to produce nutritionally enhanced rice grains via metabolic engineering, while also conferring disease resistance.
Glutelins are the most abundant seed storage proteins (SSPs) in the rice endosperm (70–80%) (Kawakatsu et al. 2008). Glutelin gene OsGluD-1 attains maximal expression levels in the starchy endosperm, commencing from 5 d after flowering (DAF) and increasing through 30 DAF (Kawakatsu et al. 2010). In this study, we employed the promoter of OsGluD-1 to drive the expression of OsNOMT, generating biofortified rice with rich sakuranetin in the endosperm. Except for the conventional liquid chromatography tandem mass spectrometry (LC–MS/MS) technique, a nondestructive, labeling-free, in situ, and visual emerging means matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) imaging was performed to detect the spatial distribution of sakuranetin and its biosynthetic pathway metabolites in pOsGluD-1::OsNOMT seeds. After observation and validation, the nutritional quality and growth of pOsGluD-1::OsNOMT were consistent with the wild type. Our work provides a successful example of biofortified crop cultivation through metabolic engineering, the biofortified rice not only enhances rice panicle blast resistance but can also be consumed as a health-promoting food and processed into dietary supplements.
Materials and Methods
Plant Material and Growth Conditions
The seeds of wild type (Oryza sativa japonica variety Zhonghua11) and transgenic lines of rice were soaked in tap water at 37 °C for 2–3 d to germination. The germinated seeds were planted in 8 × 12 cm2 boxes (individual 96-well plates) containing 1/2 MS culture solution (pH 5.8) and cultivated in a growth chamber at 30 °C, 60% relative humidity under a 12 h light/12 h dark photoperiod. Rice field in Zhejiang Academy of Agricultural Sciences (30° 18′ 44″ N, 120° 11′ 33″ E), Hangzhou, Zhejiang Province, China.
Vector Construction and Genetic Transformation
For analysis of pOsNOMT::GUS expression patterns, the genomic fragment of the OsNOMT (Os12g0240900) promoter region starting 2.5 kb upstream of the ATG codon was amplified by PCR with the primer pair pOsNOMT-F/pOsNOMT-R and sequenced. The resulting fragment was inserted into PstI and BamHI cloning sites of the expression cassette of the pCAMBIA1300-GUSplus vector to generate the plasmid pOsNOMT::GUS. For constructing p35S::OsNOMT-GFP plasmid, the full-length coding sequences of OsNOMT were amplified via RT-PCR with the primer pair OsNOMT-CDS-F1/OsNOMT-CDS-R1 and cloned into the pCAMBIA1300-35S-eGFP vector at the SpeI site. To construct the pGluD-1::OsNOMT plasmid, the OsNOMT CDS was first amplified with the primer pair OsNOMT-CDS-F2/OsNOMT-CDS-R2 and cloned into the pCAMBIA1300 vector at the PstI and HindIII sites. Subsequently, the genomic fragment of the OsGluD-1 (Os02g0249000) promoter region starting 1.6 kb upstream of the ATG codon was amplified by PCR with the primer pair pGluD-1-F/pGluD-1-R and inserted into the above construct at the KpnI and PstI sites. For generating the transgenic plants of rice, the desired constructs were transferred into Agrobacterium tumefaciens strain EHA105 as described previously (Toki et al. 2006). All transgenic plants were screened and identified by PCR amplification using universal primers (Hyg-F/Hyg-R) and the corresponding specific primers. The PCR primers can be seen in Additional file 1: Table S2.
GUS Staining
The different tissues of pOsNOMT::GUS were harvested and immersed into the staining buffer (50 mM sodium phosphate, pH 7.0, 10 mM Na2EDTA, 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide, 0.1% (v/v) Triton X-100, and 2 mM X-Gluc), vacuum-infiltrated for 5 min and incubated at 37 °C overnight. Ultimately, the tissues were cleared in 75% ethanol at 37 °C several times until the chlorophyll was completely removed. The staining patterns of GUS were observed and photographed with the digital microscope (Keyence, VHX-950F) or digital camera (Canon, 80D).
Determination of Sakuranetin and Naringenin Content
To detect the content of sakuranetin and naringenin, the different tissues of rice were harvested and ground the sample with liquid nitrogen. The sample with fresh weight of 0.1 g was weighed, and added 1.6 mL extraction buffer (the volume ratio of ethanol: water: acetonitrile: acetic acid is 79: 13.99: 7: 0.01). After 30 min of 100 Hz ultrasound, it was rotated overnight at 4 ℃. Centrifuged at 13,000 rpm for 20 min, 1 mL supernatant was absorbed. Centrifuged at 13,000 rpm for 20 min for the second time, 700 μL supernatant was absorbed into mass spectrometry tube to detected sakuranetin and naringenin by LC–MS/MS (SCIEX Triple Quad 5500+ LC–MS/MS).
RNA Extraction and qRT-PCR
Various rice tissues were frozen in liquid nitrogen and mixed with Trizol reagent. Total RNAs were extracted according to the manufacturer’s instructions (Invitrogen). cDNA was produced from 1 μg of total RNA using a reverse transcription system with gDNA Eraser (Vazyme). Quantitative real-time PCR was performed with SYBR Premix Ex TaqII (Takara) using an ABI7900HT Sequence Detection System and analyzed using the 2–△△CT method. The OsACTIN1 (Os03g0718100) was used as an internal reference to normalize the gene expression data. Three biological and two technical repeats were performed in the experiments. The qPCR primer pairs for gene amplification are given in Additional file 1: Table S2.
Sample Preparation and MALDI-MS Imaging
Rice seeds at 25 DAF were harvested and husked. The seeds were then embedded with 2% CMC in the embedding box and frozen in a dry ice-ethanol bath. After the embedding medium has fully solidified and turned white, the box can be taken out and stored at − 80 °C. Before the section, samples were placed in a − 20 °C refrigerator to equilibrate for 1 h and fixed in three drops of distilled water for cutting. The seeds were sectioned at 20 μm thickness using a Leica CM1950 cryostat (Leica Microsystems GmbH, Germany) at − 20 °C. Subsequently, the seed sections were placed in groups on electrically conductive slides coated with indium tin oxide (ITO), and dried in a vacuum desiccator for 30 min. Desiccated sections mounted on ITO glass slides were sprayed using an HTX TM sprayer (Bruker Daltonics, Germany) with 15 mg/mL DHB, dissolved in 90%: 10% acetonitrile: water. The sprayer temperature was set to 70 °C, with a flow rate of 0.1 mL/min, and a pressure of 6 psi. 28 passes of the matrix were applied to slides with 5 s of drying time between each pass.
MALDI timsTOF MS imaging experiments were performed on a prototype Bruker timsTOF flex MS system (Bruker Daltonics, Bremen, Germany) equipped with a 10 kHz smartbeam 3D laser. Laser power was set to 80% and then fixed throughout the whole experiment. The mass spectra were acquired in positive mode, and the data were obtained over a mass range from m/z (mass-to-charge ratio) 50–1300 Da. Metabolites in the samples were identified by comparing accurate m/z values, retention times (RT), and fragmentation patterns. The imaging spatial resolution was set to 50 μm, and each spectrum consisted of 400 laser shots. MALDI-MS were normalized with the Root Mean Square, and the signal intensity in each image was shown as the normalized intensity. MS/MS fragmentations performed on the timsTOF flex MS system in the MS/MS mode were used for further detailed structural confirmation of the identified metabolites.
Rice Panicle Blast Inoculation
The fungus (M. oryzae isolate Guy11) was cultured on complete medium for 2 weeks at 25 °C. The M. oryzae spores were collected in sterile water containing 0.2% Tween-20 and the concentration of spores was adjusted to about 1 × 105 spores per mL. Rice panicles at the filling stage were completely infiltrated in the spores suspension for 3 min and sealed in a plastic bag for moisturizing, and the phenotypes were observed and recorded after 5–7 days of inoculation at 28 °C. Relative fungal growth was measured by DNA-based quantitative PCR (qPCR) using the threshold cycle value (CT) of M. oryzae 28S rDNA against the CT of rice genomic ACTIN1 DNA.
Determination of Nutrition and Quality Indicators in Rice Seeds
The soluble sugar content of rice seeds was determined using the anthrone-sulphuric acid colorimetric method. Soluble sugar content was obtained by measuring the change of absorbance at 620 nm. The total amino acid content was determined based on the ninhydrin-ethanolic colorimetric method. The total amino acid content was obtained by measuring the change of absorbance at 570 nm. The total flavonoid content was determined by the aluminum-sodium nitrite colorimetric method. The total flavonoid content was obtained by measuring the change of absorbance at 510 nm. The amylose content was determined by using the iodine solution colorimetric method. The amylose content was obtained by measuring the change of absorbance at 620 nm. The total protein content is determined by the BCA method. The total protein content was obtained by measuring the change of absorbance at 562 nm. The free fatty content was measured by using the copper soap method. The free fatty content was obtained by determining the change of absorbance at 715 nm.
All these nutrition and quality indicators were determined using commercial kits (Cominbio Co., Ltd., Suzhou, China) according to the manufacturer's protocol. The absorbance of each reaction mixture was measured using a microplate reader (Infinite 200 Pro, Tecan, Switzerland).
Results
The Accumulation Pattern of Sakuranetin in Rice
Previous metabolomes found that the mature rice seeds lacked sakuranetin, but presented its biosynthetic precursor, naringenin (Shi et al. 2022; Tang et al. 2022). To confirm this result, we detected the content of naringenin through the LC–MS/MS after sampling from various tissues and developmental stages. As shown in Additional file 1: Fig. S1, we found that naringenin was high in the shoots of rice seedlings, gradually increased in roots with growth and development, and was rarely present in seeds at the filling and mature stages. To investigate the accumulation pattern of the sakuranetin in rice seeds, we initially introduced the GUS reporter gene driven by the promoter of sakuranetin biosynthetase gene OsNOMT (pOsNOMT::GUS) into rice plants to characterize the tissue-specific expression pattern of OsNOMT. The GUS-staining patterns revealed that OsNOMT was highly expressed in the leaves and leaf sheaths of rice seedlings, and was slightly expressed in the ridges and embryos of seeds at the filling stage, with no signals in roots, husks, and endosperm (Fig. 2A–C). The quantitative real-time PCR (qRT-PCR) results also showed that OsNOMT was highly expressed in the shoots of rice seedlings. The expression levels decreased gradually with the growth time, while it was almost not expressed in roots and seeds (Fig. 2D). To further analyze the accumulation pattern of sakuranetin in various rice tissues, LC–MS/MS detected the content of sakuranetin. As shown in Fig. 2E, in general agreement with the OsNOMT expression pattern and naringenin content, sakuranetin content was high in the shoots of rice seedlings and decreasing with growth and development time, whereas it was not detected in roots. However, in contrast to naringenin, sakuranetin was less abundant than naringenin in each tissue, like previous studies, was absent in mature seeds. These results indicate that sakuranetin is absent or present in rice seeds at very low abundance.
The expression pattern of OsNOMT and sakuranetin content in different rice tissues. A GUS staining of 7-day-old seedlings of pOsNOMT::GUS. Scale bar = 1 cm. B GUS staining of 20-day-old leaf (top) and leaf sheath (bottom) of pOsNOMT::GUS. Scale bar = 1 mm. C GUS staining of pOsNOMT::GUS seeds at 15 DAF. The top is the intact husked seeds, and the bottom is the cross section of the seeds (the left half contains the ridge of the seed, the right half does not). Scale bar = 0.5 mm. D qRT-PCR analysis of the expression levels of OsNOMT in different tissues of ZH11. E LC–MS/MS analysis of the sakuranetin content in different tissues of ZH11. Different letters indicate significant differences at P < 0.05 as determined by one-way ANOVA with Tukey’s test (mean ± s.d., n = 3, individual values and means are shown, biologically independent samples) (D, E). n.d., not detected
Engineering the Biosynthesis of Sakuranetin in the Rice Endosperm
Subsequently, we attempted to achieve accumulate sakuranetin in rice seeds through metabolic engineering and synthetic biology. First, we used the strong cauliflower mosaic virus 35S promoter to drive OsNOMT (p35S::OsNOMT-GFP) and transform wild-type rice ZH11 (Zhonghua11) for constitutive expression in various tissues of rice. As shown in Additional file 1: Fig. S2, while OsNOMT was successfully expressed in rice at both transcriptional and translational levels and high levels of sakuranetin could be detected in the shoots of rice (Additional file 1: Fig. S2A–C), the effect of sakuranetin accumulation in the seeds was not satisfactory (Additional file 1: Fig. S2D).
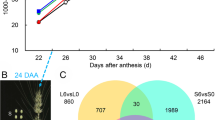
Endosperm-specific SSP gene (e.g., GluA, GluB, GluC, GluD, and Gt) promoters have been widely utilized in plant molecular farming, where the expression levels of the OsGluD-1 gradually increase at least until the rice seeds mature stage 30 DAF, in contrast to the gradual decrease of many other glutelin genes at later stages of seed development (Kawakatsu et al. 2008; He et al. 2011; Patti et al. 2012). Therefore, we employed 1.6 kb OsGluD-1 promoter to drive the CDS of OsNOMT (pGluD-1::OsNOMT) to express OsNOMT in the rice endosperm strongly (Fig. 3A). As expected, the expression levels of OsNOMT (Additional file 1: Fig. S3A) and the content of sakuranetin (Fig. 3B) in rice seeds at the filling stage were found to be notably higher than wild type in three transgenic lines by qRT-PCR and LC–MS/MS. This value is higher than the sakuranetin content in the shoots of p35S::OsNOMT-GFP plants (Additional file 1: Fig. S2C), and even higher than sakuranetin induced by rice blast-infected in wild-type rice (Hasegawa et al. 2014). Surprisingly, the naringenin content was also increased in the pGluD-1::OsNOMT plants (Additional file 1: Fig. S3B), probably due to feedback regulation of the metabolic pathway. MALDI-MS imaging is a powerful technique developed in recent years for detecting the spatial distribution of metabolites and has been widely used in plant and food fields. Previous studies have used MALDI-MS imaging to reveal the distribution of different metabolites in many fruits and seeds, such as tomato fruits, strawberry fruits, Brassica napus seeds, and maize seeds (Feenstra et al. 2017; Lu et al. 2018; Wang et al. 2021; Li et al. 2022). To get a better insight into observing the accumulation pattern of sakuranetin in pGluD-1::OsNOMT seeds, we performed MALDI-MS imaging to detect the spatial distribution of relevant metabolites in the sakuranetin biosynthesis pathway in seeds at the dough stage. As shown in Fig. 3C and Additional file 1: Table S1, the levels of sakuranetin, naringenin/naringenin chalcone, and cinnamic acid were elevated in pGluD-1::OsNOMT plants, with unchanged levels of the synthesis initiator L-phenylalanine, and decreased levels of dihydrokaempferol. This is consistent with the previous LC–MS/MS results. Moreover, the spatial distribution of these metabolites was inconsistent. L-phenylalanine, naringenin/naringenin chalcone, and sakuranetin were almost uniformly distributed throughout the seeds, cinnamic acid was mainly distributed in the embryo and seed cortex, and dihydrokaempferol was mainly distributed in the endosperm. In the pGluD-1::OsNOMT plants, sakuranetin was increased throughout the seed, and dihydrokaempferol was decreased mainly in the endosperm. Notably, the spatial distribution of sakuranetin in WT is slightly different from the expression pattern of OsNOMT (Fig. 2C). The OsNOMT predominately expressed in the embryo and the dorsal side of rice seed, whereas sakuranetin accumulated throughout the seed. There are two possible reasons for this phenomenon. First, the samples of GUS staining were whole or half WT seeds, and OsNOMT gene expression patterns could be detected. However, the accumulation of sakuranetin in WT seeds is extremely low (Fig. 2E), and the thickness of the spatial metabolome sample section is only 20 μm, so the spatial distribution of sakuranetin cannot be accurately and meticulously detected by MALDI-MS imaging. Secondly, rice seeds may not be sliced precisely to the ridges of the seed, such as in the last image in Fig. 2C. Therefore, GUS staining and MALDI-MS imaging have different sensitivities. These two approaches complement each other, providing evidence for modifications of metabolic pathways from upstream gene levels and downstream metabolite levels, respectively.
The specific expression of OsNOMT in endosperm resulted in the accumulation of sakuranetin and increased the blast resistance in rice seeds. A Schematic diagram of the pOsGluD-1::OsNOMT construct with the OsNOMT CDS, driven by the OsGluD-1 promoter. B LC–MS/MS analysis of the sakuranetin content in ZH11 and pOsGluD-1::OsNOMT seeds at 15 DAF. #6, #7, and #17 represent three different transgenic lines. P-values were determined using two-tailed Student’s t-tests, ***P < 0.001 (mean ± s.d., n = 3, individual values and means are shown, biologically independent samples). FW, fresh weight. C The MALDI-MS images of metabolites in the sakuranetin biosynthesis pathway in ZH11 and pOsGluD-1::OsNOMT seeds at 25 DAF. The colors in the spatial distribution images represent the relative content of metabolites, and 0 to 1 indicates sequentially increasing metabolite content. Scale bar = 1 mm. D LC–MS/MS analysis of the sakuranetin content in ZH11 and pOsGluD-1::OsNOMT seeds at the mature stage. DW, dry weight; n.d., not detected. E The phenotype of pOsGluD-1::OsNOMT panicle inoculated with rice blast. The red triangles indicate M. oryzae-infected rice seeds. Scale bar = 2 cm. F The relative fungal growth in E. Relative fungal growth indicates the relative fungal biomass in M. oryzae-inoculated panicles and was determined with fungal 28S rDNA normalized to rice genomic ACTIN1 DNA by DNA-based qPCR. P-values were determined using two-tailed Student’s t-tests, ***P < 0.001 (mean ± s.d., n = 3, individual values and means are shown, biologically independent samples)
Finally, we detected sakuranetin in rice seeds at the mature stage, and pGluD-1::OsNOMT plants were still much higher than the wild type. Still, it was lower than the filling stage (Fig. 3D). Considering the strong activity of sakuranetin against rice blast, the rice blast resistance of pGluD-1::OsNOMT panicles was determined. Once rice seeds are infected with the M. oryzae, the episperm will show disease spots or even turn black and die. As shown in Fig. 3E, the panicle of pGluD-1::OsNOMT had more seeds infected with M. oryzae than the wild type. Further detection of the relative fungal growth by DNA-based qPCR revealed that the M. oryzae biomass of transgenic panicles was much less than wild type (Fig. 3F). These suggested that the panicle blast resistance of pGluD-1::OsNOMT was significantly higher than that of the wild type. Generally, these results indicated that endosperm-specific expression of OsNOMT successfully increased the sakuranetin content and the rice blast resistance of rice seeds.
The Nutrition and Quality of pOsGluD-1::OsNOMT Seeds Were not Affected
To gain insights into whether our modification to the endosperm affect rice seed nutrition and quality, we comprehensively analyzed the contents of total amino acids, soluble sugars, total flavonoids, amylose, total protein, and free fatty acids in transgenic rice and wild type.
As the basic unit of protein, amino acids participate in not only redox homeostasis, osmoregulation, signal transduction, and other life processes but also the content of some amino acids such as aspartic acid and glutamic acid in rice grains, can affect the eating quality (Matsuzaki et al. 1992; Kelly and Pearce 2020; Guo et al. 2021). The MALDI-MS imaging detected 9 essential amino acids (L-methionine, L-isoleucine, L-leucine, L-valine, L-lysine, L-phenylalanine, L-tryptophan, L-threonine, and L-histidine) and 2 amino acids that determine the eating quality (L-aspartic acid and L-glutamic acid) of rice seeds at the dough stage (Figs. 3C, 4A and Additional file 1: Table S1). It is found that the L-histidine content in pOsGluD-1::OsNOMT seeds increased, and the main increasing segmentation was the embryo, while the content of L-methionine and L-isoleucine/L-leucine slightly decreased. Their distribution fell in the embryo and raised in the endosperm. The contents and spatial distribution of other amino acids did not change significantly. Furthermore, the content of total amino acids in the mature seeds of transgenic plants showed no significant difference from the wild type, suggesting that changes in the content and spatial distribution of a few amino acids did not affect the content of total amino acids (Fig. 4C). Soluble sugars are important energy storage substances and the main source of sweetness in plant seeds. Their content is closely related to the nutrition and quality of seeds (Zhu et al. 2007). As shown in Fig. 4B and Additional file 1: Table S1, the detection of the 4 main soluble sugars (glucose, fructose, sucrose, and maltose) by MALDI-MS imaging revealed that the soluble sugar content was slightly elevated in pOsGluD-1::OsNOMT seeds, and the main segmentation of the elevation was in the embryo. We also analyzed the total soluble sugar content of transgenic plant seeds at the mature stage and observed no significant difference compared to the wild type, which may be due to the other soluble sugars in the rice seeds (Fig. 4D). In addition to amino acids and soluble sugars, the content of flavonoids, amylose, protein, and free fatty acids in seeds is also a crucial factor in determining its nutrition, taste and quality (Birla et al. 2017; Kasote et al. 2022). The contents of total flavonoid, amylose, total protein and free fatty acid in the mature seeds were detected, and there was no significant difference between pOsGluD-1::OsNOMT plants and wild type (Fig. 4E–H). In summary, these results show that the nutrition and quality of pOsGluD-1::OsNOMT seeds were not affected.
Comparison of nutrition and quality between pOsGluD-1::OsNOMT and wild type seeds. A The MALDI-MS images of the essential amino acids and amino acids that determine the eating quality in ZH11 and pOsGluD-1::OsNOMT seeds at 25 DAF. Scale bar = 1 mm. B The MALDI-MS images of of targeted free soluble sugars in ZH11 and pOsGluD-1::OsNOMT seeds at 25 DAF. Scale bar = 1 mm. C–H Comparison of total amino acid, soluble sugar, total flavonoid, amylose, total protein and free fatty acid content between ZH11 and pOsGluD-1::OsNOMT seeds at the mature stage. P-values were determined using two-tailed Student’s t-tests (mean ± s.d., n = 3, individual values and means are shown, biologically independent samples)
The Growth and Development of pOsGluD-1::OsNOMT Plants Were not Affected
Apparently, the use of metabolic engineering to increase the biosynthesis of sakuranetin in endosperm caused changes in the content of some metabolites (Figs. 3C, 4A, B). Next, we explored whether this modification affected the growth and development of pOsGluD-1::OsNOMT plants. We tracked and observed the phenotypes of pOsGluD-1::OsNOMT plants at different growth stages, and found that the plant architecture, panicle morphology, and maturity stage were the same as the wild type (Fig. 5A–C). As a key agricultural trait, grain size determines grain yield and quality in rice (Zuo and Li 2014). The grain size of pOsGluD-1::OsNOMT plants was recorded and analyzed in focus, and there were no significant differences between the three transgenic lines and the wild type in terms of grain width (Fig. 5D, G) and grain length (Fig. 5E, H). The threshed seeds also showed the same phenotype as the wild type (Fig. 5F). In addition, we measured the 1,000-grain weight of transgenic plants, and there was no significant difference between pOsGluD-1::OsNOMT plants and wild type (Fig. 5I). Altogether, these results demonstrated that the growth and development of pOsGluD-1::OsNOMT were not affected. In fact, based on our observations in the phytotron and the field, we also found the vegetative and reproductive phenotypes of p35S::OsNOMT-GFP were not significantly different from the WT at all stages of growth and development. This suggested that the accumulation of sakuranetin in various tissues of rice does not influence its growth and development.
The phenotype of pOsGluD-1::OsNOMT. A Phenotype of 14-day-old ZH11 and pOsGluD-1::OsNOMT seedlings. Scale bar = 5 cm. B Phenotypes of ZH11 and pOsGluD-1::OsNOMT at the reproductive stage. Scale bar = 15 cm. C The phenotypes of panicles at the grain maturation stage detached from the ZH11 and pOsGluD-1::OsNOMT. Scale bar = 3 cm. D, E Mature grains of ZH11 and pOsGluD-1::OsNOMT. Scale bar = 0.5 cm. F Husked grains of ZH11 and pOsGluD-1::OsNOMT. Scale bar = 0.8 cm. G–I Comparison of grain width (n = 10), grain length (n = 10), and 1000-grain weight (n = 3) between ZH11 and pOsGluD-1::OsNOMT. P values were determined using two-tailed Student’s t-tests (mean ± s.d., individual values and means are shown, biologically independent samples)
The Proposed Work Model of pOsGluD-1::OsNOMT
To investigate whether this work is generalizable to other cereal crops, we examined naringenin and sakuranetin content in maize (Zea mays), sorghum (Sorghum bicolor), millet (Setaria italica), oat (Avena sativa), and coix (Semen coicis). As shown in Additional file 1: Fig. S4, naringenin and sakuranetin were much higher in maize and sorghum than in the other grains, while the naringenin and sakuranetin contents in millet, oat, and coix were essentially similar to the rice. Overall, these cereal seeds lacked sakuranetin or contained very little sakuranetin. Thus, our strategy of metabolic engineering to accumulate sakuranetin in endosperm can be extended to other cereal crops.
Based on all the above results, a working model of pOsGluD-1::OsNOMT in rice seeds was proposed (Fig. 6). Specifically, the expression of the sakuranetin biosynthesis gene OsNOMT was driven by the promoter of the endosperm-specific glutelin gene OsGluD-1, resulting in the successful accumulation of sakuranetin in rice seeds. Sakuranetin content was highest at the filling stage and decreased with seed maturation. During the filling stage, the rapid and massive accumulation of sakuranetin is proposed to improve disease resistance in crop panicles by acting as a phytoalexin. At the mature stage, the lower abundance of sakuranetin confers potential nutritional and health benefits to humans. It should be noted that this modification does not affect the nutrition and yield of rice. Therefore, our study represents an entirely new paradigm for generating biofortified crops through metabolic engineering and synthetic biology, a win–win strategy for both plants and humans.
The proposed work model illustrating how endosperm-specific expression of OsNOMT leads to the accumulation of sakuranetin in the rice seeds. The expression of the sakuranetin biosynthesis gene OsNOMT was driven by the promoter of the endosperm-specific glutelin gene OsGluD-1, and OsNOMT successfully catalyzes naringenin synthesis of sakuranetin in the rice seeds. Sakuranetin content was highest at the filling stage and decreased with seed maturation. During the filling stage, the rapid and massive accumulation of sakuranetin is proposed to improve disease resistance in crop panicles by acting as a phytoalexin. At the mature stage, the lower abundance of sakuranetin confer potential nutritional and health benefits to humans. The dark brown color in the proposed model represents the rice seed embryo, the yellow color represents the cortex (rice bran layer), the beige color represents the endosperm, and the small red dots represent sakuranetin
Discussion
Plants have evolved a variety of defense mechanisms against pathogen attack, such as strengthening local cell walls, regulating phytohormone signals, activating plant innate immune response, and synthesizing phytoalexins (López et al. 2008; Piasecka et al. 2015; Hacquard et al. 2017). Rice phytoalexins include diterpenoids, phenylamides, and flavonoids, of which sakuranetin is the only flavonoid phytoalexin (Valletta et al. 2023). In fact, compared with sakuranetin and uncommon phenylamides, the research on diterpenoid phytoalexins is more extensive and in-depth. However, there are numerous species of diterpenoids phytoalexin, and the synthetic pathway is multistep and complex (Schmelz et al. 2014; Morelli and Rodriguez-Concepcion 2023). Moreover, the enzymes involved in the synthesis of diterpenoids phytoalexin have different catalytic mechanisms, different tissue specificity in plant distribution, and different suborganelle localization in the plastid, which increases the difficulty of modifying their metabolic pathways (Lange and Ghassemian 2003). In addition to phytoalexin, glucosinolates (GSLs) are another well-known class of secondary defense metabolites produced by plants, which mainly accumulate in the seeds of Brassicales plants (Grubb and Abel 2006; Mitreiter and Gigolashvili 2021). However, being an anti-nutritional factor, it can significantly impinge on the taste and nutritional value of the plant's edible tissues. As a result, the use of metabolic engineering strategies aimed at enriching rice seeds with glucosinolates would not be considered an optimal choice either (Xu et al. 2023). In recent years, the strong activity against rice leaf blast of sakuranetin has been revealed by many studies (Kodama et al. 1992; Hasegawa et al. 2014; Murata et al. 2020). Relatively, with a simple structure, clear synthetic steps, a lack of odor, and diverse physiological activities, sakuranetin presents a promising solution for balancing resistance, growth, nutrition, and quality in rice. Seed is an important organ of rice that provides energy for plant growth and nutrients for humans. Nevertheless, sakuranetin is deficient in the rice grains (Fig. 2E). In this study, the sakuranetin-rich biofortified rice was generated by modifying the biosynthesis pathway of rice endosperm. As expected, the panicle blast resistance of the transgene rice was significantly higher than WT (Fig. 3E, F).
It is widely recognized that the metabolites in plants are characterized by species specificity and spatio-temporal specificity, meaning that certain metabolic processes and enzymes responsible for catalyzing these metabolic processes exhibit the same properties (Hanson and Gregory 2002; De-la-Cruz Chacón et al. 2013). The isoenzymes distributed in various tissues may give rise to different catalytic properties and regulatory mechanisms. Furthermore, the substrate levels, coenzymes, and other factors related to enzyme activity or regulatory mechanisms are also diverse in various tissues. Therefore, modifying of a specific metabolic pathway in one plant is often challenging to apply to another species. For instance, the activities of dihydrofavonol 4-reductase (DFR) and anthocyanidin synthase (ANS) are required in tissues such as leaves, fruits, and corollas of many dicot plants that naturally synthesize anthocyanins (Wang et al. 2022). In contrast, the endosperm of the monocot plants rice is inherently deficient in anthocyanins, which complicates the modification of their synthesis pathway in the rice endosperm (Zhu et al. 2017). Unlike anthocyanins, sakuranetin, which is also flavonoid, is naturally present in rice, even though its content and the expression of the synthase gene OsNOMT vary in different tissues and developmental stages of rice (Fig. 2D, E). We achieved efficient and massive biosynthesis of sakuranetin in the rice endosperm by introducing a single-step metabolic pathway using rice endogenous promoters, synthesis genes and substrates. A precedent exists for utilizing species' endogenous advantages to produce biofortified crops easily and efficiently. Li et al. knocked out the enzyme gene that catalyzes 7-DHC (a precursor for provitamin D3 biosynthesis) biosynthesis of cholesterol in Solanaceae plants by genome editing and produced a provitamin D3-rich biofortified plant by modifying a single-step metabolic pathway (Li et al. 2022).
Besides being essential for plants to resist various stresses, sakuranetin has many health-promoting effects (Stompor 2020; Junaid et al. 2022). For example, sakuranetin exhibits anti-tumor, anti-viral, anti-parasitic, anti-inflammatory, and antioxidant physiological activities. Previous studies have found that sakuranetin demonstrates anti-proliferative properties against ESCC, B16BL6 melanoma, and Colo 320 (Ugocsai et al. 2005; Hong and Ying 2015; Drira and Sakamoto 2016). Sakuranetin was also reported to have antiviral activity against human rhinovirus 3 and influenza B virus (Choi 2017; Kwon et al. 2018). In addition, sakuranetin attenuated chronic allergic airway inflammation in mice by inhibiting MAPK and STAT3-SOCS3 (Santana et al. 2019). Rice is a vital food crop with a long history of cultivation and consumption. It provides more than 20% of the calories for more than half of the world's population and up to 50% of the calories for the population of Asia (Gutaker et al. 2020; Sapwarobol et al. 2021). Consequently, the endogenous synthesis of sakuranetin in rice seeds is of great significance to human health. Biofortified rice can not only supply people with sakuranetin conveniently and directly in their daily diet. Still, it can also be developed and processed into healthcare products to satisfy a broader consumption range.
In pOsGluD-1::OsNOMT plants, the sakuranetin levels were observed to plummet as the rice reached maturity (Fig. 3B, D). According to Kawakatsu et al., the expression levels of OsGluD-1 remained high in the late stage of seed development (Kawakatsu et al. 2008), which ruled out the possibility that the work efficiency of pOsGluD-1::OsNOMT dropped sharply at the mature stage. The substrate naringenin content was reduced in seeds at the mature stage (Additional file 1: Fig. S1), which may partially explain the reduced sakuranetin content. Ogo et al. once specifically co-expressed OsPAL and OsCHS by OsGluB-1 promoter in rice endosperm to achieve naringenin accumulation (Ogo et al. 2013). Therefore, simultaneously specifically expressing flavonoid synthase genes upstream of OsNOMT in the endosperm to generate a sufficient supply of naringenin could result in higher levels of sakuranetin in rice seeds. We utilized OsGluD-1, which is more persistently expressed than OsGluB-1 in rice seeds, as an endosperm-specific promoter to achieve sakuranetin accumulation in seeds efficiently. Sakuranetin not only has the nutritional function of naringenin and other flavonoids but is also unique for its brilliant resistance to phytopathogen.
Moreover, based on our detection in various cereal crops, naringenin is prevalent in the grain. At the same time, sakuranetin is deficient or insufficient (Additional file 1: Fig. S4), suggesting that our modification strategy has the potential to be applied to other cereal crops. In recent years, the emerging MALDI-MS imaging technique has been used to investigate the dynamic changes of amino acids during germination of rice seeds and the spatial distribution of lipids in mature rice seeds (Ren et al. 2023; Zhang et al. 2023). With the MALDI-MS imaging, we not only revealed the spatial distribution of sakuranetin for the first time but also evaluated the spatial distribution of multiple nutrients in the seeds of pOsGluD-1::OsNOMT plants (Figs. 3C, 4A, B). Before this study, HPLC and DPBA (diphenyl boric acid 2-amino ethyl ester) staining were commonly used to detect flavonoids in biofortified rice seeds (Ogo et al. 2013, 2016). Herein, more specifical, precise, and comprehensive observation on the spatial distribution of multiple metabolites in different segmentations of rice seeds provides a new assessment method for detecting the effects of metabolic engineering.
Conclusion
Collectively, we generated a sakuranetin-rich biofortified rice by metabolic engineering in which the expression of the sakuranetin biosynthesis gene OsNOMT was driven by the promoter of the endosperm-specific glutelin gene OsGluD-1, resulting in the successful accumulation of sakuranetin in rice seeds. The seeds of pOsGluD-1::OsNOMT plants may combine both resistance and nutritional dual functions, and this modification of the metabolic pathway did not affect plant nutrition and growth. This innovative attempt at engineering sakuranetin-rich endosperm rice would motivate the nutrient fortification of cereal crops.
Availability of Data and Materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- NOMT:
-
Naringenin 7-O-methyltransferase
- LC–MS/MS:
-
Liquid chromatography tandem mass spectrometry
- MALDI-MS:
-
Matrix-assisted laser desorption/ionization mass spectrometry
- BPH:
-
Brown planthopper
- ESCC:
-
Esophageal squamous cell carcinoma
- Colo 320:
-
Colon cancer
- PAL:
-
Phenylalanine ammonialyase
- C4H:
-
Cinnamic acid 4-hydrolase
- 4CL:
-
Coumarin-CoA ligase
- CHS:
-
Chalcone synthase
- CHI:
-
Isomerase
- SSPs:
-
Seed storage proteins
- DAF:
-
Days after flowering
- ZH11:
-
Zhonghua11
- GSLs:
-
Glucosinolates
- DFR:
-
Dihydrofavonol 4-reductase
- ANS:
-
Anthocyanidin synthase
- DPBA:
-
Diphenyl boric acid 2-amino ethyl ester
References
Birla DS, Malik K, Sainger M, Chaudhary D, Jaiwal R, Jaiwal PK (2017) Progress and challenges in improving the nutritional quality of rice (Oryza sativa L.). Crit Rev Food Sci Nutr 57:2455–2481
Butelli E, Licciardello C, Zhang Y, Liu J, Mackay S, Bailey P, Reforgiato-Recupero G, Martin C (2012) Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24:1242–1255
Chiu LW, Zhou X, Burke S, Wu X, Prior RL, Li L (2010) The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol 154:1470–1480
Choi HJ (2017) In vitro antiviral activity of sakuranetin against human rhinovirus 3. Osong Public Health Res Perspect 8:415–420
De-la-Cruz Chacón I, Riley-Saldaña CA, González-Esquinca AR (2013) Secondary metabolites during early development in plants. Phytochem Rev 12:47–64
Drira R, Sakamoto K (2016) Sakuranetin induces melanogenesis in B16BL6 melanoma cells through inhibition of ERK and PI3K/AKT signaling pathways. Phytother Res 30:997–1002
Espley RV, Brendolise C, Chagné D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens RP, Allan AC (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21:168–183
Feenstra AD, Alexander LE, Song Z, Korte AR, Yandeau-Nelson MD, Nikolau BJ, Lee YJ (2017) Spatial mapping and profiling of metabolite distributions during germination. Plant Physiol 174:2532–2548
Grubb CD, Abel S (2006) Glucosinolate metabolism and its control. Trends Plant Sci 11:89–100
Guo N, Zhang S, Gu M, Xu G (2021) Function, transport, and regulation of amino acids: what is missing in rice? Crop J 9:530–542
Gutaker RM, Groen SC, Bellis ES, Choi JY, Pires IS, Bocinsky RK, Slayton ER, Wilkins O, Castillo CC, Negrão S, Oliveira MM, Fuller DQ, Guedes JAD, Lasky JR, Purugganan MD (2020) Genomic history and ecology of the geographic spread of rice. Nat Plants 6:492–502
Ha SH, Kim JK, Jeong YS, You MK, Lim SH, Kim JK (2019) Stepwise pathway engineering to the biosynthesis of zeaxanthin, astaxanthin and capsanthin in rice endosperm. Metab Eng 52:178–189
Hacquard S, Spaepen S, Garrido-Oter R, Schulze-Lefert P (2017) Interplay between innate immunity and the plant microbiota. Annu Rev Phytopathol 55:565–589
Hanson AD, Gregory JF (2002) Synthesis and turnover of folates in plants. Curr Opin Plant Biol 5:244–249
Hasegawa M, Mitsuhara I, Seo S, Okada K, Yamane H, Iwai T, Ohashi Y (2014) Analysis on blast fungus-responsive characters of a flavonoid phytoalexin sakuranetin; accumulation in infected rice leaves, antifungal activity and detoxification by fungus. Molecules 19:11404–11418
He Y, Ning T, Xie T, Qiu Q, Zhang L, Sun Y, Jiang D, Fu K, Yin F, Zhang W, Shen L, Wang H, Li J, Lin Q, Sun Y, Li H, Zhu Y, Yang D (2011) Large-scale production of functional human serum albumin from transgenic rice seeds. Proc Natl Acad Sci U S A 108:19078–19083
He Y, Zhao Y, Hu J, Wang L, Li L, Zhang X, Zhou Z, Chen L, Wang H, Wang J, Hong G (2024) The OsBZR1-OsSPX1/2 module fine-tunes the growth-immunity trade-off in adaptation to phosphate availability in rice. Mol Plant 17:258–276
Hong L, Ying SH (2015) Ethanol extract and isolated constituents from Artemisia dracunculus inhibit esophageal squamous cell carcinoma and induce apoptotic cell death. Drug Res 65:101–106
Huang D, Wang X, Tang Z, Yuan Y, Xu Y, He J, Jiang X, Peng SA, Li L, Butelli E, Deng X, Xu Q (2018) Subfunctionalization of the Ruby2-Ruby1 gene cluster during the domestication of citrus. Nat Plants 4:930–941
Junaid M, Basak B, Akter Y, Afrose SS, Nahrin A, Emran R, Shahinozzaman M, Tawata S (2022) Sakuranetin and its therapeutic potentials—a comprehensive review. Z Naturforsch C J Biosci 78:27–48
Kasote D, Sreenivasulu N, Acuin C, Regina A (2022) Enhancing health benefits of milled rice: current status and future perspectives. Crit Rev Food Sci Nutr 62:8099–8119
Kawakatsu T, Yamamoto MP, Hirose S, Yano M, Takaiwa F (2008) Characterization of a new rice glutelin gene GluD-1 expressed in the starchy endosperm. J Exp Bot 59:4233–4245
Kawakatsu T, Hirose S, Yasuda H, Takaiwa F (2010) Reducing rice seed storage protein accumulation leads to changes in nutrient quality and storage organelle formation. Plant Physiol 154:1842–1854
Kelly B, Pearce EL (2020) Amino assets: how amino acids support immunity. Cell Metab 32:154–175
Kodama O, Miyakawa J, Akatsuka T, Kiyosawa S (1992) Sakuranetin, a flavanone phytoalexin from ultraviolet-irradiated rice leaves. Phytochemistry 31:3807–3809
Kwon DH, Ji JH, Yim SH, Kim BS, Choi HJ (2018) Suppression of influenza B virus replication by sakuranetin and mode of its action. Phytother Res 32:2475–2479
Lange BM, Ghassemian M (2003) Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol Biol 51:925–948
Li J, Scarano A, Gonzalez NM, D’Orso F, Yue Y, Nemeth K, Saalbach G, Hill L, de Oliveira MC, Moran R, Santino A, Martin C (2022) Biofortified tomatoes provide a new route to vitamin D sufficiency. Nat Plants 8:611–616
Liu M, Hong G, Li H, Bing X, Chen Y, Jing X, Gershenzon J, Lou Y, Baldwin IT, Li R (2023) Sakuranetin protects rice from brown planthopper attack by depleting its beneficial endosymbionts. Proc Natl Acad Sci U S A 120:e2305007120
López MA, Bannenberg G, Castresana C (2008) Controlling hormone signaling is a plant and pathogen challenge for growth and survival. Curr Opin Plant Biol 11:420–427
Lu S, Sturtevant D, Aziz M, Jin C, Li Q, Chapman KD, Guo L (2018) Spatial analysis of lipid metabolites and expressed genes reveals tissue-specific heterogeneity of lipid metabolism in high- and low-oil Brassica napus L. seeds. Plant J 94:915–932
Mano H, Ogasawara F, Sato K, Higo H, Minobe Y (2007) Isolation of a regulatory gene of anthocyanin biosynthesis in tuberous roots of purple-fleshed sweet potato. Plant Physiol 143:1252–1268
Matsuzaki A, Takano T, Sakamoto S, Kuboyama T (1992) Relation between eating quality and chemical components in milled rice and amino acid contents in cooked rice. Jpn J Crop Sci 61:561–567
Melo MNO, Oliveira AP, Wiecikowski AF, Carvalho RS, Castro JL, de Oliveira FAG, Pereira HMG, da Veiga VF, Capella MMA, Rocha L, Holandino C (2018) Phenolic compounds from Viscum album tinctures enhanced antitumor activity in melanoma murine cancer cells. Saudi Pharm J 26:311–322
Mitreiter S, Gigolashvili T (2021) Regulation of glucosinolate biosynthesis. J Exp Bot 72:70–91
Morelli L, Rodriguez-Concepcion M (2023) Open avenues for carotenoid biofortification of plant tissues. Plant Commun 4:100466
Murata K, Kitano T, Yoshimoto R, Takata R, Ube N, Ueno K, Ueno M, Yabuta Y, Teraishi M, Holland CK, Jander G, Okumoto Y, Mori N, Ishihara A (2020) Natural variation in the expression and catalytic activity of a naringenin 7-O-methyltransferase influences antifungal defenses in diverse rice cultivars. Plant J 101:1103–1117
Ogawa S, Miyamoto K, Nemoto K, Sawasaki T, Yamane H, Nojiri H, Okada K (2017) OsMYC2, an essential factor for JA-inductive sakuranetin production in rice, interacts with MYC2-like proteins that enhance its transactivation ability. Sci Rep 7:40175
Ogo Y, Ozawa K, Ishimaru T, Murayama T, Takaiwa F (2013) Transgenic rice seed synthesizing diverse flavonoids at high levels: a new platform for flavonoid production with associated health benefits. Plant Biotechnol J 11:734–746
Ogo Y, Mori T, Nakabayashi R, Saito K, Takaiwa F (2016) Transgenic rice seed expressing flavonoid biosynthetic genes accumulate glycosylated and/or acylated flavonoids in protein bodies. J Exp Bot 67:95–106
Paine JA, Shipton CA, Chaggar S, Howells RM, Kennedy MJ, Vernon G, Wright SY, Hinchliffe E, Adams JL, Silverstone AL, Drake R (2005) Improving the nutritional value of golden rice through increased pro-vitamin A content. Nat Biotechnol 23:482–487
Park HL, Yoo Y, Hahn TR, Bhoo SH, Lee SW, Cho MH (2014) Antimicrobial activity of UV-induced phenylamides from rice leaves. Molecules 19:18139–18151
Patti T, Bembi B, Cristin P, Mazzarol F, Secco E, Pappalardo C, Musetti R, Martinuzzi M, Versolatto S, Cariati R, Dardis A, Marchetti S (2012) Endosperm-specific expression of human acid beta-glucosidase in a waxy rice. Rice 5:34
Piasecka A, Jedrzejczak-Rey N, Bednarek P (2015) Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol 206:948–964
Rakwal R, Agrawal GK, Yonekura M, Kodama O (2000) Naringenin 7-O-methyltransferase involved in the biosynthesis of the flavanone phytoalexin sakuranetin from rice (Oryza sativa L.). Plant Sci 155:213–221
Ren Z, Qin L, Chen L, Xu H, Liu H, Guo H, Li J, Yang C, Hu H, Wu R, Zhou Y, Xue K, Liu B, Wang X (2023) Spatial Lipidomics of EPSPS and PAT transgenic and non-transgenic soybean seeds using matrix-assisted laser desorption/ionization mass spectrometry imaging. J Agric Food Chem 71:10190–10202
Santana FPR, da Silva RC, Grecco SDS, Pinheiro AJMCR, Caperuto LC, Arantes-Costa FM, Claudio SR, Yoshizaki K, Macchione M, Ribeiro DA, Tibério IFLC, Lima-Neto LG, Lago JHG, Prado CM (2019) Inhibition of MAPK and STAT3-SOCS3 by sakuranetin attenuated chronic allergic airway inflammation in mice. Mediat Inflamm 2019:1356356
Sapwarobol S, Saphyakhajorn W, Astina J (2021) Biological functions and activities of rice bran as a functional ingredient: a review. Nutr Metab Insights 14:11786388211058560
Schmelz EA, Huffaker A, Sims JW, Christensen SA, Lu X, Okada K, Peters RJ (2014) Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J 79:659–678
Shi Y, Guo Y, Wang Y, Li M, Li K, Liu X, Fang C, Luo J (2022) Metabolomic analysis reveals nutritional diversity among three staple crops and three fruits. Foods 11:550
Shimizu T, Lin F, Hasegawa M, Okada K, Nojiri H, Yamane H (2012a) Purification and identification of naringenin 7-O-methyltransferase, a key enzyme in biosynthesis of flavonoid phytoalexin sakuranetin in rice. J Biol Chem 287:19315–19325
Shimizu T, Lin F, Hasegawa M, Nojiri H, Yamane H, Okada K (2012b) The potential bioproduction of the pharmaceutical agent sakuranetin, a flavonoid phytoalexin in rice. Bioengineered 3:352–357
Stompor M (2020) A review on sources and pharmacological aspects of sakuranetin. Nutrients 12:513
Sun C, Deng L, Du M, Zhao J, Chen Q, Huang T, Jiang H, Li CB, Li C (2020) A transcriptional network promotes anthocyanin biosynthesis in tomato flesh. Mol Plant 13:42–58
Tang J, Li X, Zhang Y, Yang Y, Sun R, Li Y, Gao J, Han Y (2022) Differential flavonoids and carotenoids profiles in grains of six poaceae crops. Foods 11:2068
Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47:969–976
Ugocsai K, Varga A, Molnár P, Antus S, Molnár J (2005) Effects of selected flavonoids and carotenoids on drug accumulation and apoptosis induction in multidrug-resistant colon cancer cells expressing MDR1/LRP. In Vivo 19:433–438
Valletta A, Iozia LM, Fattorini L, Leonelli F (2023) Rice phytoalexins: half a century of amazing discoveries; part I: distribution, biosynthesis, chemical synthesis, and biological activities. Plants 12:260
Wang J, Yang E, Chaurand P, Raghavan V (2021) Visualizing the distribution of strawberry plant metabolites at different maturity stages by MALDI-TOF imaging mass spectrometry. Food Chem 345:128838
Wang L, Chen M, Lam PY, Dini-Andreote F, Dai L, Wei Z (2022) Multifaceted roles of flavonoids mediating plant-microbe interactions. Microbiome 10:233
Xu D, Sanden NCH, Hansen LL, Belew ZM, Madsen SR, Meyer L, Jørgensen ME, Hunziker P, Veres D, Crocoll C, Schulz A, Nour-Eldin HH, Halkier BA (2023) Export of defensive glucosinolates is key for their accumulation in seeds. Nature 617:132–138
Ye X, Al-Babili S, Klöti A, Zhang J, Lucca P, Beyer P, Potrykus I (2000) Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287:303–305
Zhang Y, Zhang Y, Shi Y (2023) A reliable and effective sample preparation protocol of MALDI-TOF-MSI for lipids imaging analysis in hard and dry cereals. Food Chem 398:133911
Zhu C, Cheng C, Liu X, He J (2007) Accumulation of soluble sugars related to desiccation tolerance during rice (Oryza sativa L) seed development and maturation. Seed Sci Technol 11:649–659
Zhu Q, Yu S, Zeng D, Liu H, Wang H, Yang Z, Xie X, Shen R, Tan J, Li H, Zhao X, Zhang Q, Chen Y, Guo J, Chen L, Liu YG (2017) Development of “Purple Endosperm Rice” by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency transgene stacking system. Mol Plant 10:918–929
Zhu Q, Zeng D, Yu S, Cui C, Li J, Li H, Chen J, Zhang R, Zhao X, Chen L, Liu YG (2018) From Golden Rice to aSTARice: bioengineering astaxanthin biosynthesis in rice endosperm. Mol Plant 11:1440–1448
Zhu Q, Tan J, Liu YG (2022) Molecular farming using transgenic rice endosperm. Trends Biotechnol 40:1248–1260
Zuo J, Li J (2014) Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu Rev Genet 48:99–118
Acknowledgements
We thank Dr. Hongxuan Lin (CAS Centre for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, CAS) for the donation of pCAMBIA1300-GUSplus plasmid. We thank Metware Biotechnology Co., Ltd. (Wuhan, China) for technical support in MALDI-MS imaging.
Funding
This work was supported by the Zhejiang Provincial Natural Science Foundation of China (LR22C020003 and LQ23C020003), the National Natural Science Foundation of China (32272553), and the State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products.
Author information
Authors and Affiliations
Contributions
Y.Z. and G.H. designed the project; Y.Z., J.H., Z.Z., L.L., and J.W. prepared materials and performed the experiments; Y.Z. and G.H. performed the data analysis and wrote the manuscript; L.L., X.Z., Y.H., and C.Z. revised the manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
All authors have agreed for publication.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1. Fig. S1.
LC-MS/MS analysis of the naringenin content in different tissues of ZH11. Fig. S2. The sakuranetin content in p35S::OsNOMT-GFP. Fig. S3. The expression level of OsNOMT and naringenin content in pOsGluD-1::OsNOMT seeds at the filling stage. Fig. S4. The naringenin content and sakuranetin content in different cereal crops. Table S1. The relative quantification of selected metabolites by MALDI-MS imaging. Table S2. Primers list.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhao, Y., Hu, J., Zhou, Z. et al. Biofortified Rice Provides Rich Sakuranetin in Endosperm. Rice 17, 19 (2024). https://doi.org/10.1186/s12284-024-00697-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-024-00697-w