Abstract
Background
With the increasing frequency of climatic anomalies, high temperatures and long-term rain often occur during the rice-harvesting period, especially for early rice crops in tropical and subtropical regions. Seed dormancy directly affects the resistance to pre-harvest sprouting (PHS). Therefore, in order to increase rice production, it is critical to enhance seed dormancy and avoid yield losses to PHS. The elucidation and utilization of the seed dormancy regulation mechanism is of great significance to rice production. Preliminary results indicated that the OsMKKK62-OsMKK3-OsMPK7/14 module might regulate ABA sensitivity and then control seed dormancy. The detailed mechanism is still unclear.
Results
The overexpression of OsMKK3 resulted in serious PHS. The expression levels of OsMKK3 and OsMPK7 were upregulated by ABA and GA at germination stage. OsMKK3 and OsMPK7 are both located in the nucleus and cytoplasm. The dormancy level of double knockout mutant mkk3/mft2 was lower than that of mkk3, indicating that OsMFT2 functions in the downstream of MKK3 cascade in regulating rice seeds germination. Biochemical results showed that OsMPK7 interacted with multiple core ABA signaling components according to yeast two-hybrid screening and luciferase complementation experiments, suggesting that MKK3 cascade regulates ABA signaling by modulating the core ABA signaling components. Moreover, the ABA response and ABA responsive genes of mpk7/14 were significantly higher than those of wild-type ZH11 when subjected to ABA treatment.
Conclusion
MKK3 cascade mediates the negative feedback loop of ABA signal through the interaction between OsMPK7 and core ABA signaling components in rice.
Similar content being viewed by others
Background
Seed dormancy inhibits seed germination under adverse environment conditions and increases the survival and fitness of wild plants. For rice, dormancy affects the consistency of seedling emergence and the resistance to pre-harvest sprouting (PHS). The consistency of seedling emergence affects the yield, and the resistance to PHS affects the final harvest yield and quality, both of which are important links in crop production (Shu et al. 2016). For rice production, moderate levels of seed dormancy are favorable. Dormancy is inadvertently lost in the long-term breeding process. Uncovering the mechanisms driving dormancy regulation is critical for rice production and yield optimization, particularly by enhancing moderate dormancy in seeds. ABA and GA are the primary hormones controlling seed dormancy, and the ABA/GA balance determines dormancy or germination. The ABA signal promotes the production and maintenance of seed dormancy (Shu et al. 2016; Kucera et al. 2005). ABA signaling is determined by ABA content and ABA sensitivity. ABA content is mainly controlled by NCED (9-cis-epoxycarotenoid dioxygenase), catalyzing the limiting steps of ABA synthesis, and ABA8OX (ABA8'-hydroxylase, also known as Cytochrome P450 Monooxygenase, CYP707A), which regulates the limiting steps in degradation (Lefebvre et al. 2006). ABA sensitivity is mainly related to the generation mechanism of ABA signal. PYR/PYL/RCAR-PP2C-SnRK2, the generation pathway of an ABA signal, is widely accepted (Fujii et al., 2009). TaMFT has been proved to control seed dormancy (Liu et al. 2015; Nakamura et al. 2011), which interacts with OsbZIP23/66/72 and regulates the downstream ABA responsive genes (Song et al. 2020). OsMFT2, the homolog of TaMFT in rice, was negatively regulated by MKK3 cascade (Mao et al. 2019). Further investigation is necessary in order to clarify the regulatory mechanism of the MKK3 cascade on seed dormancy.
The MAPK cascade is an evolutionarily conserved signaling module that plays diverse roles in plants (Rodriguez et al. 2010), which magnifies the signal through sequential phosphorylation of downstream proteins, integrating the information between the surrounding environment and the metabolic response centers. OsMKK3 was induced by mechanical wounding, infestation of Xanthomonas oryzae, and treatments with methyl jasmonate or salicylic acid, suggesting that it could play diverse roles in different conditions (Jalmi and Sinha 2016; Zhou et al. 2019; Sözen et al. 2020). Phenotype analysis results also showed that OsMKK3/OrMKK3 influenced seedling growth, grain size, and eating quality (Pan et al. 2021a, b). MKK3 is effectively a multi-signal distribution and switching center. It can mediate ABA, hydrogen peroxide and light signals, which may be involved in dormancy regulation (Dóczi et al. 2007; Sethi et al. 2014; Danquah et al. 2015). Multiple MAPK genes have been proved to regulate seed dormancy, which expanded our understanding in dormancy research. MKK1–MAPK6 positively affects seed dormancy and the expression of AtCAT1 in Arabidopsis (Xing et al. 2009, 2008). In Arabidopsis, two MKKK genes, AtRAF10 and AtRAF11, controlled seed dormancy by regulating the expression of AtABI3 and AtABI5 (Lee et al. 2015), which were key genes in ABA response (Du et al. 2018; Hobo et al. 1999; Zou et al. 2008).
Overexpression of OsMKKK62 resulted in the loss of dormancy in rice. Knockouts of the downstream members, OsMKK3, OsMPK7, and OsMPK14, lead to the evident increase of seed dormancy level, indicating that MKK3 cascade might substantially influence ABA signaling (Mao et al. 2019). According to sequence similarity, AtMKKK14/15/16/17/18-AtMKK3-AtMPK1/2/7/14 should function like OsMKKK62-OsMKK3-OsMPK7/14 (MKK3 cascade) (Rao et al. 2010; Hamel et al. 2006). ABA induced the expression of AtMKKK18 and promoted stomatal closure (Mitula et al. 2015). PP2C protein, ABI1, interacted with At MKKK18 and promoted its degradation (Mitula et al. 2015). One group also reported the interaction between ABI1 and AtMKKK18 (Choi et al. 2017). Another finding demonstrated the activation of AtMPK7 induced by ABA was inhibited in the mutants, atmkk3-1 and atmkkk17/18 (Danquah et al. 2015). These results indicated that the MKK3 cascade might take part in the ABA signal. The homolog gene of AtMKK3 controlled seed dormancy in barley and wheat (Nakamura et al. 2016; Torada et al. 2016). In rice, the MKK3 cascade negatively regulated ABA sensitivity and seed dormancy (Mao et al. 2019). In Arabidopsis, similar phenomena have been observed (Danquah et al. 2015; Mitula et al. 2015; Choi et al. 2017), although the regulatory effect in Arabidopsis thaliana was smaller than that in rice, suggesting that the MKK3 cascade influences ABA signaling greatly.
In this study, we explored the expression characteristics of MKK3 cascade members, checked the germination phenotype of related mutants, identified several OsMPK7 interaction proteins, and finally proposed that MKK3 cascade mediated the negative feedback loop of ABA signaling in rice.
Results
Overexpression of OsMKK3 Resulted in PHS and Decreased the Tolerance to Dehydration
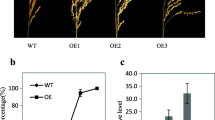
The overexpression of AtMKK3 did not lead to vivipary in Arabidopsis thaliana (Danquah et al. 2015). To further check the function of OsMKK3, we created OsMKK3-overexpressing (OsMKK3OE) transgenic lines. OsMKK3 was under the control of the maize ubiquitin promoter (Ubi:OsMKK3). By checking the hygromycin resistance at germination stage, three homozygous lines (OsMKK3OE1, OsMKK3OE2, OsMKK3OE3) were selected for characterization. The gene expression analysis results showed that the transcript levels of OsMKK3 were higher in leaves of the three OsMKK3OE lines than that in ZH11 at three-leaf stage (Fig. 1A). To test the germination character, we harvested the panicles and treated them under germination conditions for two days. All seeds from overexpression lines germinated, but no wild-type ZH11 seeds germinated (Fig. 1B). At 25 days after heading, small cracks appeared in the embryonic part of OsMKK3OE husk (Fig. 1C), indicating that the germination process began before full maturity even without rain. A germination experiment showed that OsMKK3OE seeds could germinate more quickly than wild-type ZH11 seeds (Fig. 1D). To evaluate the influence of dehydration tolerance, we carried out the germination experiment after drying (50 °C, three days). The germination rate of OsMKK3OE seeds decreased substantially, while the wild-type ZH11 germinated normally (Fig. 1E), suggesting that dehydration tolerance decreased greatly in OsMKK3OE seeds.
Overexpression of OsMKK3 leads to PHS and the loss of dehydration tolerance. A Relative expression level of OsMKK3 in leaves of OsMKK3OE lines. Values represent the mean ± SD of three biological replicates. Student’s t-tests were used to generate p values. (** = p < 0.01) B Germination phenotype of the OsMKK3OE panicles. After 25 days of heading, panicles of overexpression lines and ZH11 were sampled for germination tests, and the photos were taken after two days of treatment. C After 25 days of heading, small cracks appeared in the embryonic part of OsMKK3OE husk (Scale bar = 2 mm). D Freshly harvested seeds were used for germination test, and germination percentage was calculated at the indicated time. Values represent the mean ± SD of three biological replicates (30 seeds for each replicate). E The germination rates of the dry seeds were calculated after four days germination treatment. Values represent the mean ± SD of three biological replicates (30 seeds for each replicate). Student’s t-tests were used to generate p values. (** = p < 0.01)
Organizational Expression and Subcellular Localization and of OsMKKK62, OsMKK3 and OsMPK7
To investigate the biological role of OsMKK3 in rice, we analyzed the spatial and temporal expression of OsMKK3 in different rice tissues. OsMKK3 was ubiquitously expressed in the whole plant, and the transcription levels were relatively high in the shoot of the three-leaf stage, the first leaf and the young panicle before heading (Fig. 2A). We also analyzed the expression of OsMPK7 and OsMKKK62 with a similar method, showing that OsMPK7 was also widely expressed in the whole plant, the transcription levels were relatively high in young panicles and shoots and OsMKKK62 was mainly expressed in root and stem (Fig. 2A). The subcellular localization of OsMKK3 was determined through transient expression of an OsMKK3-GFP fusion protein in rice protoplasts. Fluorescence from the fusion protein was observed in the nuclei and cytoplasm (Fig. 2B), indicating that OsMKK3-GFP was located in both. To further learn the function of the MKK3 cascade, we checked the subcellular localization of OsMPK7 and OsMKKK62. With the same method, OsMPK7-GFP and OsMKKK62-GFP did not show a clear signal. To further check the subcellular localization of OsMPK7, we constructed another fusion vector, GFP-OsMPK7, introduced it in the protoplasts, and observed the bright fluorescence signal at nuclei and cytoplasm (Fig. 2B). These results indicated that MKK3 cascades may function in both nuclei and cytoplasm.
Expression characteristics of OsMKKK62, OsMKK3 and OsMPK7. A Transcription analysis of OsMKKK62, OsMKK3 and OsMPK7 in different tissues by quantitative RT-PCR. Shoot and root were sampled at three-leaf stage; young panicle, first leaf, stem and mature root were sampled before heading; embryos were sampled at the late period of dough stage. Values represent the means ± SD of three biological replicates. B Subcellular localization of OsMKK3 and OsMPK7 in rice protoplasts. GFP, OsMKK3-GFP and GFP-OsMPK7 driven by the 35S promoter under green fluorescence, red fluorescence, bright field, and merged views (Scale bar = 10 μm). C, D The expression of OsMKK3 (C) and OsMPK7 (D) in embryos treated with ABA or GA. The seeds were treated with 100 μM ABA, 100 μM GA and water respectively. After 3, 6, 12, and 24 h of treatment, ten embryos were cut for gene expression analysis. Values represent the mean ± SD of three biological replicates. Student’s t-tests were used to generate p values. (* = p < 0.05, ** = p < 0.01)
ABA and GA Induced the Expression of OsMKK3 and OsMPK7
ABA and GA are the primary hormones controlling seed dormancy. To identify the induction effect of ABA and GA on MKK3 cascades, we treated the seeds with ABA and GA solution, and performed expression analysis with the embryos. The results showed that both ABA and GA could induce the expression of OsMKK3 and OsMPK7, the induction effect of ABA lasted for a long time, and the induction effect of GA disappeared quickly (Fig. 2C, D). The induction effect of ABA or GA on OsMKKK62 is not obvious (Data not shown). These results indicated that MKK3 cascade may be involved in the signal transmission of both ABA and GA.
Hydrogen Peroxide Could Promote the Germination of mpk7/14
Biochemical experimental results showed that the MKK3 cascade may be involved in the signal of hydrogen peroxide in Arabidopsis (Dóczi et al. 2007). Hydrogen peroxide is a molecular hub of ROS signal and often acts as a rice germination promoter (Chen et al. 2016; Peng et al. 2022). We tested the germination of mpk7/14with exogenous 20 mM hydrogen peroxide. In order to eliminate the effect of residual hydrogen peroxide on germination, all seeds were dehusked. The results indicated that hydrogen peroxide could obviously promote the germination of mpk7/14 (Fig. 3).
Germination result of mpk7/14in hydrogen peroxide. Before germination test, all seeds were dehusked. The seeds were treated with 20 mM hydrogen peroxide and water as control. At the indicated time the germinated seeds were counted. Values represent the mean ± SD of three biological replicates (30 seeds for each replicate)
Knockout of OsMFT2 Could Partially Rescue the Germination Phenotype of mkk3 Mutant
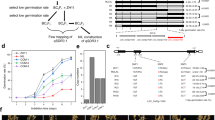
Expression analysis showed that OsMFT2 was negatively regulated by MKK3 cascade (Mao et al. 2019). AtMFT, the homologous gene of OsMFT2 in Arabidopsis, was a vital gene in the ABA signal pathway (Xi et al. 2010). To check the functional relationship between the MKK3 cascade and OsMFT2, we constructed CRISPR vector to edit OsMFT2 and OsMKK3 simultaneously, and introduced the vector into ZH11 through Agrobacterium-mediated genetic transformation. According to the sequencing chromatogram of the T2 generation (Additional file 1: Fig. S1), four independent Crispr lines were screened for planting (Fig. 4A) and the transgenic seeds were dehusked for a germination test. MKK3/mft2 (3 nucleotide deletion in OsMKK3, 1 nucleotide insertion in OsMFT2) showed a germination phenotype similar to that of wild-type ZH11. The mutation of OsMKK3 inhibited seed germination (Mao, et al. 2019). OsMFT2 negatively regulated seed germination (Song et al. 2020). The germination performance of MKK3/mft2 indicated that the 3-nucleotide deletion in OsMKK3 did not affect its function. The germination performance of MKK3/mft2 could represent the phenotype of the osmft2 mutant. When OsMKK3 and OsMFT2 were knocked out simultaneously (mkk3/mft2-1, mkk3/mft2-2, mkk3/mft2-3), the germination rates were significantly higher than that of mkk3 (Fig. 4B). The result suggested that the mutation of OsMFT2 could partially rescue the germination phenotype of osmkk3 mutant, and MKK3 cascade possibility acted on the upstream of OsMFT2 in the dormancy regulation pathway.
Germination phenotype of Crispr lines. A Schematic of four Crispr lines. Red letters indicate the exon sequences, blue letters indicate the intron sequences, green letters indicate the inserted nucleotide and black letters indicate amino acids. Dashs indicate the deleted nucleotides. Asterisks indicate termination codons. PAM, protospacer adjacent motif. Red triangles indicate the target position. B Germination phenotype of OsMKK3/OsMFT2 Crispr lines. The germination rates were calculated at 12 h intervals. Values represent the mean ± SD of three biological replicates (30 dehusked seeds for each replicate)
Identification of Interaction Proteins of OsMPK7
ABA is the primary hormone that regulates seed dormancy and germination, which regulates ABA responsive genes through core ABA signaling components (PYR/PYL/RCAR-PP2C-SnRKs) (Cutler et al. 2010; Hubbard et al. 2010). In Arabidopsis, Snrk2.2/2.3/2.6 play vital roles in ABA signal transduction and the triple mutant exhibited a loss of seed dormancy (Nakashima et al. 2009; Fujii and Zhu 2009). As the homologs of Snrk2.2/2.3/2.6 in rice, OsSAPK8/9/10 were also activated by ABA, suggesting their functions in dormancy regulation (Kobayashi et al. 2004). Among these three genes, the expression level of OsSAPK8 is the highest in later stages of maturation (Sato et al. 2013), suggesting its crucial function in seed dormancy. We were interested in whether OsMPK7 could interact with OsSAPK8 (Fig. 5A). To test this, we performed the Y2H experiment. According to the growth on QD, OsMPK7 did not interact with OsSAPK8. With the same method, we checked interaction relationships of OsMPK7 with 10 OsPLYs and 3 OsPP2Cs. The results showed that OsMPK7 could interact with 6 OsPLYs and OsPP2C50 (Fig. 5A). To further confirm these interactions, we selected OsPLY7, OsPLY11 and OsPP2C50 to perform Luciferase Complementation experiments. Chemiluminescence imaging results also supported the interaction of OsMPK7 with these three proteins (Fig. 5B–D). All the interaction results indicated that the MKK3 cascade regulated ABA signal through the interaction between OsMPK7 and core ABA signaling components.
Interaction analysis between OsMPK7 and core ABA signaling components. A Y2H assay of interactions between OsMPK7 and ABA signaling precursors. QD (Quadruple-dropout medium lacking Ade, His, Leu and Trp); DD, (Double-dropout medium lacking Leu and Trp). The numbers above represent the cell concentration (cells/mL), and the proteins expressed in the yeast are listed on the left. B–D Luciferase complementation results of OsMPK7’s interaction with OsPLY7, OsPLY11 and OsPP2C50 in N. benthamiana leaves
ABA-Response was Upregulated in mpk7/14
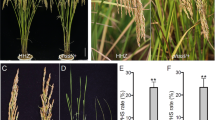
A batch of interactions between OsMPK7 and the core ABA signaling component indicated the close relationship between the MKK3 cascade and the ABA signal. To evaluate the effect of ABA on mpk7/14 mutant, mpk7/14 seeds were treated with ABA for six days. Under mock treatment (0 μM ABA), there was no significant difference in germination rate between ZH11 and mpk7/14, but under 0.2 μM ABA, the germination rates of mpk7/14 seeds (15.6% and 17.8%) were extremely significantly lower than that of ZH11 (70%) (Fig. 6A). Under 2 μM ABA, the germination rate of ZH11 was 42.2%, and all mpk7/14 seeds could not germinate (Fig. 6A, B). Moreover, the shoot lengths and root lengths of ZH11 were significantly longer than those of mpk7/14 without or with ABA treatment (Fig. 6C). The root lengths of ZH11, mpk7/14–2 and mpk7/14–3 under 0.2 μM ABA treatment were 43.0%, 23.8% and 17.2% of those under mock treatment, respectively (Additional file 2: Fig. S2). The inhibition effect of 0.2 μM ABA on mpk7/14 was significantly stronger than that on ZH11, suggesting that mpk7/14 was more sensitive to ABA than that of ZH11.
Mutant mpk7/14 showed enhanced ABA response. A The germination performance of ZH11 and mpk7/14. The germinated seeds were counted after 6 days of ABA treatment, Values represent the mean ± SD of three biological replicates (30 seeds for each replicate). The asterisks indicate significant differences compared with ZH11. Student’s t-tests were used to generate p values (*p < 0.05, **p < 0.01). B Phenotypes of ZH11and mpk7/14 after 6 days of ABA treatment on 1/2 MS culture medium without or with ABA (0.2 μM or 2 μM) (Scale bar = 1 cm). C The lengths of shoots and roots of ZH11 and mpk7/14 after 6 days of ABA treatment. Values represent the mean ± SD of three biological replicates (10 germinated seeds for each replicate). The asterisks indicate significant differences compared with mock treatment. Student’s t-tests were used to generate p values (**p < 0.01). D Expression analysis of ABA responsive genes in ZH11 and mpk7/14. Seeds were soaked in water for six hours at 30 °C and ten embryos were cut for gene expression analysis. Values represent the mean ± SD of three biological replicates. The asterisks indicate significant differences compared with ZH11. Student’s t-tests were used to generate p values (*p < 0.05, **p < 0.01)
To further check this deduction, we selected several ABA responsive genes (Tian et al. 2015; Zang et al. 2016; Mukherjee et al. 2006; Wang et al. 2022) and compared their expression levels in mpk7/14 and wild-type ZH11. The results showed that the expression levels of ABA responsive genes in mpk7/14 were significantly higher than those in ZH11 (Fig. 6D), suggesting that MKK3 cascade negatively regulates ABA signal.
Discussion
The consistency of germination affects seedling raising, and the resistance to PHS affects the quality of harvested seeds, both of which are important for crop production. Seed dormancy negatively regulates consistent germination and positively regulates PHS tolerance. Moderate seed dormancy is suitable for crop production.
OsMKKK62 and LOC103652526 (the maize homolog of OsMKKK62) positively regulate the germination of rice (Mao et al. 2019), indicating MKK3 cascade plays an important role in seed germination in Gramineae. The overexpressed OsMKK3 resulted in PHS and further confirmed that the MKK3 cascade negatively regulated seed dormancy (Fig. 1B). Overexpression of AtMKK3 showed little effect on germination in Arabidopsis (Danquah et al. 2015). This suggests stark differences in the dormancy regulation mechanism between rice and Arabidopsis.
The results from Arabidopsis showed that the AtMKKK17/18, the homologous gene of OsMKKK62, was induced by ABA in leaves (Matsuoka et al. 2015; Mitula et al. 2015; Danquah et al. 2015). Our results showed that ABA and GA could induce the expression of OsMKK3 and OsMPK7 in germinating seeds (Fig. 2C, D). ABA and GA antagonistically regulate seed germination (Shu et al. 2016). MKK3 cascade positively regulates germination (Fig. 1B) (Mao et al. 2019). There should be an interesting mechanism governing this contradictory phenomenon among ABA, GA, and MKK3 cascades. The expression of AtMFT is regulated by ABA (Xi et al. 2010). OsMFT2 is the rice homologous gene of AtMFT and the transcription of OsMFT2 is regulated by MKK3 cascade (Mao et al. 2019). Knockout experiments showed that the germination rate of double mutant (mkk3/mft2-1, mkk3/mft2-2 and mkk3/mft2-3) was faster than that of mkk3 (Fig. 4B). These results indicated that MKK3 cascade may function upstream of OsMFT2. Both ABA and GA could regulate the expression of AtMFT (Xi et al. 2010). SLR1 encodes a DELLA protein, which is a key negative regulator in GA signaling. SLR1 mutant didn’t show serious PHS phenotype (Ikeda et al. 2001), indicating that the effect of GA on germination was not as strong as MKK3 cascade. Snrk2.2/2.3/2.6 positively regulated ABA signal and the triple mutant showed vivipary phenotype (Nakashima et al. 2009; Fujii and Zhu 2009), which is similar with the PHS phenotype of OsMKK3OE (Fig. 1A). It indicated that MKK3 cascade may regulate seed dormancy mainly through ABA signal. ABA receptors interact with PP2C, releasing SAPK from PP2C repression and producing the ABA signal, which results in the regulation of abiotic stress resistance and growth development including germination (Park et al. 2009; Cutler et al. 2010; Klingler et al. 2010). There are 13 PLYs in rice (He et al. 2014; Yadav et al. 2020; Miao et al. 2018). We cloned 10 OsPLYs in a prey vector. Through Y2H experiments, we proved that that OsMPK7 interacted with 7 OsPLYs. In addition, we checked the interaction relationship of OsMPK7 with 3 OsPP2Cs by Y2H. The results showed that OsMPK7 interacted with OsPP2C50 (Fig. 5A). The LUC experiment also supported the interactions of OsMPK7 with OsPYL6, OsPYL11 and OsPP2C50 (Fig. 5B–D). Results of ABA treatment and expression analysis of ABA responsive genes also suggested that MKK3 cascade negatively regulated ABA signals (Figs. 6, Additional file 2: Fig. S2). We speculated that OsMPK7 could accept the recruitment of OsPYLs and OsPP2C, and then restrain the operation of ABA core components from producing ABA signals, thus promoting germination (Fig. 7). Recently, AtMKK3-AtMPK7-AtERF4-AtEXPA module was confirmed to regulate seed germination in Arabidopsis thaliana (Chen et al. 2023). Further experiments are needed to determine whether there is a similar regulatory pathway in rice.
Working model of MKK3 cascade integrating core ABA signaling. Green frames indicate positive regulatory factors of ABA signals, and red frames indicate negative regulatory factors. Arrows indicate positive regulation; flat-ended arrows indicate negative regulation; double lines indicate protein interactions
Some PLYs existed in monomer state, produced ABA signals in the absence of ABA and ensured a basal level of ABA signaling in plant growth, while other PLYs existed in dimer state and produce ABA signal by combining molecular ABA (Hao et al. 2011; Tian et al. 2015; Dupeux et al. 2011). OsMPK7 could interact with dimer OsPYL1 or monomer OsPYL6, suggesting MKK3 cascade could regulate basal ABA signaling and the ABA signals induced by molecular ABA. OsMPK7 and OsMKK3 were induced by ABA and inhibited the ABA signals, suggesting that MKK3 cascade was in the negative feedback loop of ABA signaling (Fig. 7). In addition, GA and hydrogen peroxide could induce the expression of OsMKK3 and OsMPK7 (Fig. 2C, D) (Jalmi and Sinha 2016), and hydrogen peroxide could partially rescued the germination phenotype of mpk7/14, so the MKK3 cascade may mediate the crosstalk among ABA, GA and hydrogen peroxide signals.
OsPP2C68 was specifically localized in the nucleus of rice protoplasts, but OsPP2C50 and OsPP2C6 were not (Min et al. 2019). The subcellular localization corresponded to the clades observed in phylogenetic trees (Min et al. 2019; Kim et al. 2012). OsPYL1-10 were localized in the nucleus and cytosol of Nicotiana benthamiana leaves (Tian et al. 2015). OsMPK7 was localized in the nucleus and cytosol (Fig. 2B), and interacted with OsPLY1, OsPLY4, OsPLY5, OsPLY6, OsPLY7, OsPLY9, OsPLY11 and OsPP2C50 (Fig. 6). These results suggest that the interaction between OsMPK7 and core ABA signaling components cannot be inferred from the results of subcellular localization or simple evolutionary similarity, and that MKK3 cascades may be regulating ABA signal through fine regulation of parts of core ABA signaling components, which may be formed by later evolution.
Hydrogen peroxide is a popular germination promoter (Chen et al. 2016; Peng et al. 2022). In Arabidopsis, hydrogen peroxide could activate AtMPK7 in an AtMKK3-mediated manner (Dóczi et al. 2007). In rice, hydrogen peroxide could upregulate the expression of OsMPK7 (Jalmi and Sinha 2016). The positive regulation effect of MKK3 cascade on rice germination process was obvious (Fig. 1B) (Mao et al. 2019). Therefore, hydrogen peroxide was likely to play a regulatory role through MKK3 cascade. When treated with exogenous hydrogen peroxide treatment, the germination of mpk7/14 was partially rescued (Fig. 3), suggesting that MKK3 cascade may not be the only pathway of hydrogen peroxide signal in germination. ABA could promote the production of hydrogen peroxide in guard cells (Pei et al. 2000; Wang and Song 2008). In the process of germination, the relationship between ABA and hydrogen peroxide needs further research.
Conclusion
Our results demonstrated that MKK3 cascade mediated the negative feedback regulation of ABA signal. In the plant lifecycle, stresses and favorable conditions often occur randomly. ABA signals promote adaption to stress condition, and the MKK3 cascade may be always ready to inhibit the ABA signal and start normal growth. Both of these are beneficial for plants to quickly adapt to stressful environment or develop in favorable conditions. Based on the strong effects on ABA signaling in growth and development, MKK3 cascade may be a major participant in the negative feedback loop of ABA signaling and more related studies should be performed to reveal its working mechanism.
Methods
Plant Materials and Growth Conditions
All rice (Oryza sativa) plants used in this study were the rice cultivar ZH11 or the transgenic progeny of ZH11. The OsMKK3 knockout line (mkk3-1 and mkk3-3) and the OsMPK7/14 knockout line (mpk7/14-2 and mpk7/14-3) were created by gene edition (Mao et al. 2019).
Development of Overexpression Plants and Crispr Plants
For construction of the OsMKK3-overexpression vector, the OsMKK3 coding sequence was amplified with cDNA from ZH11 as template and inserted into a pOX vector at the HindIII site by recombinant cloning (the inserted gene was controlled by ubiquitin promoter). The primers used in this paper are listed in Additional file 3: Table S1. For construction of the CRISPR vector, the target sites were designed on the web (http://cbi.hzau.edu.cn/crispr/). Two target sites were selected, which were allocated in the first exon of OsMKK3 and the third exon of OsMFT2, respectively. The CRISPR vector was created with guidance from the literature (Ma et al. 2015). The overexpression vector and the CRISPR vector were introduced into ZH11 by Agrobacterium tumefaciens (EHA105)-mediated transformation. The subsequent selection of overexpression plants and Crispr plants were performed as described in previous study (Mao et al. 2019).
Yeast Two-Hybrid Assay
OsMPK7 CDS was inserted into pGBKT7 vector as the bait. The cDNA obtained from ZH11 was used as the template to amplify OsSAPK8, OsPLYs and OsPP2Cs with specific primers. All the PCR products were inserted into the pGADT7 as preys, respectively. Yeast Two-Hybrid experiments were conducted with OsMPK7-bait and performed as described in previous study (Mao et al. 2019).
Luciferase Complementation Assay
The fragment of OsMPK7 CDS was inserted into pCAMBIA1300-nLUC vector, and the OsPLYs and OsPP2C50 CDS were inserted into the pCAMBIA1300-cLUC vector, and then introduced into Agrobacterium tumefaciens GV3101. Agrobacterium containing pCAMBIA1300-nLUC-OsMPK7 was co-injected with each Agrobacterium containing cLUC fusion vector into the same part of the N. benthamiana leaf and incubated for 48 h. After D-luciferin treatment, LUC activity was photographed using chemiluminescence imaging (FUSION FX EDGE SPECTRA).
Germination Test
The newly harvested panicles were immediately soaked in water for 5 min, drained of water briefly, and kept in 30 °C and humidity > 95%. The germination phenotype was checked two days later.
Thirty seeds were distributed on a filter paper in a 9-cm dish, soaked with 10 mL water, and kept in an incubator (30 °C, humidity > 95%). The presence of 1 mm protrusions in the embryo was used as the standard of germination, and germinated seeds were counted every 12 h.
Subcellular Localization
For protein subcellular localization analysis, the coding sequences of OsMKK3 and OsMPK7 were amplified using primers listed in Additional file 3: Table S1 and inserted into the GFP vector to produce OsMKK3-GFP and GFP-OsMPK7 fusion constructs. Subsequently, the fusion constructs were transformed into rice stem protoplasts as described previously (Zhang et al. 2011). After incubation in the dark for 24 h at room temperature, GFP fluorescence was detected by laser confocal microscopy (Zeiss LSM710, Germany).
Gene Expression Analysis
In the seed treatment, high-quality ZH11 seeds were placed in a 9 cm petri dish containing 10 ml working solution. Working solution was either 100 μm ABA or 100 μM GA, with water as control. The dishes were kept in a 30 °C incubator. Ten embryos were cut at 3, 6, 12, and 24 h of treatment and stored in liquid nitrogen until RNA extraction. Total RNA was extracted from plant tissue with Trizol reagent (Invitrogen) according to the manufacturer’s instructions. Quantitative RT-PCR analysis was performed as described in a previous report (Mao et al. 2019).
ABA Treatment
Thirty seeds were dehusked and sterilized with 75% ethanol for 30 s and 2.5% sodium hypochlorite solution for 25 min, washed three times with water and then placed on 1/2 MS culture medium without or with ABA (0.2 μM or 2 μM) for 6 days in a 27 °C constant temperature incubator.
Accession Numbers
Sequence data from this article can be found in the Rice Genome Annotation Project or NCBI Database under the following accession numbers:
OsMKKK62 (LOC_Os01g50420); OsMKK3 (LOC_Os06g27890); OsMPK7 (LOC_Os06g48590); OsMFT2(LOC_Os01g02120); OsSAPK8(LOC_Os03g55600); OsABA45(LOC_Os12g29400); OsLEA3(LOC_Os05g46480); OsLip9(LOC_Os02g44870); OsbZIP5 (LOC_Os01g46970); OsbZIP23(LOC_Os02g52780); OsRAB16A(LOC_Os11g26790); OsRAB16B(LOC_Os11g26780); OsPYL1(LOC_Os01g61210); OsPYL2(LOC_Os02g13330); OsPYL3(LOC_Os02g15640); OsPYL4(LOC_Os03g18600); OsPYL5(LOC_Os05g12260); OsPYL6(LOC_Os05g39580); OsPYL7(LOC_Os06g33640); OsPYL9(LOC_Os06g36670); OsPYL10(LOC_Os10g42280); OsPYL11(Os6g0526400); OsPP2C6(LOC_Os01g40094); OsPP2C50(LOC_Os05g46040); OsPP2C68(LOC_Os09G15670).
Availability of Data and Materials
The datasets supporting the conclusions of this article are provided within the article and its additional files.
Abbreviations
- PHS:
-
Pre-harvest sprouting
- ABA:
-
Abscisic acid
- GA:
-
Gibberellin
- Y2H:
-
Yeast two-hybrid
- QD:
-
Quadruple-dropout medium lacking Ade, His, Leu and Trp
- MKK3 cascade:
-
OsMKKK62-OsMKK3-OsMPK7/14
References
Chen BX, Li WY, Gao YT, Chen ZJ, Zhang WN, Liu QJ, Chen Z, Liu J (2016) Involvement of polyamine oxidase-produced hydrogen peroxide during coleorhiza-limited germination of rice seeds. Front Plant Sci 7:1219
Chen X, Li Q, Ding L, Zhang S, Shan S, Xiong X, Jiang W, Zhao B, Zhang L, Luo Y, Lian Y, Kong X, Ding X, Zhang J, Li C, Soppe WJJ, Xiang Y (2023) The MKK3–MPK7 cascade phosphorylates ERF4 and promotes its rapid degradation to release seed dormancy in Arabidopsis. Mol Plant 16(11):1743–1758
Choi S, Lee S, Na Y, Jeung S, Kim SY (2017) Arabidopsis MAP3K16 and other salt-inducible MAP3Ks regulate ABA response redundantly. Mol Cells 40(3):230–242
Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61:651–679
Danquah A et al (2015) Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J 82(2):232–244
Dóczi R, Brader G, Pettkó-Szandtner A, Rajh I, Djamei A, Pitzschke A, Teige M, Hirt H (2007) The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. Plant Cell 19(10):3266–3279
Du L, Xu F, Fang J, Gao S, Tang J, Fang S, Wang H, Tong H, Zhang F, Chu J, Wang G, Chu C (2018) Endosperm sugar accumulation caused by mutation of PHS8/ISA1 leads to pre-harvest sprouting in rice. Plant J 95(3):545–556
Dupeux F, Santiago J, Betz K, Twycross J, Park SY, Rodriguez L, Gonzalez-Guzman M, Jensen MR, Krasnogor N, Blackledge M, Holdsworth M, Cutler SR, Rodriguez PL, Márquez JA (2011) A thermodynamic switch modulates abscisic acid receptor sensitivity. EMBO J 30(20):4171–4184
Fujii H, Zhu JK (2009) Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci 106(20):8380–8385
Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462(7273):660–664
Hamel LP, Nicole MC, Sritubtim S, Morency MJ, Ellis M, Ehlting J, Beaudoin N, Barbazuk B, Klessig D, Lee J, Martin G, Mundy J, Ohashi Y, Scheel D, Sheen J, Xing T, Zhang S, Seguin A, Ellis BE (2006) Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci 11(4):192–198
Hao Q, Yin P, Li W, Wang L, Yan C, Lin Z, Wu JZ, Wang J, Yan SF, Yan N (2011) The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins. Mol Cell 42(5):662–672
He Y, Hao Q, Li W, Yan C, Yan N, Yin P (2014) Identification and characterization of ABA receptors in Oryza sativa. PLoS ONE 9(4):e95246–e95246
Hobo T, Kowyama Y, Hattori T (1999) A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc Natl Acad Sci 96(26):15348–15353
Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI (2010) Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev 24(16):1695–1708
Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13(5):999–1010
Jalmi SK, Sinha AK (2016) Functional involvement of a mitogen activated protein kinase module, OsMKK3-OsMPK7-OsWRK30 in mediating resistance against Xanthomonas oryzae in rice. Sci Rep 6:37974
Kim H, Hwang H, Hong JW, Lee YN, Ahn IP, Yoon IS, Yoo SD, Lee S, Lee SC, Kim BG (2012) A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J Exp Bot 63(2):1013–1024
Klingler JP, Batelli G, Zhu JK (2010) ABA receptors: the START of a new paradigm in phytohormone signalling. J Exp Bot 61(12):3199–3210
Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T (2004) Differential activation of the rice sucrose nonfermenting1–related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 16(5):1163
Kucera B, Cohn MA, Leubner-Metzger G (2005) Plant hormone interactions during seed dormancy release and germination. Seed Sci Res 15(4):281–307
Lee SJ, Lee MH, Kim JI, Kim SY (2015) Arabidopsis putative MAP kinase kinase kinases Raf10 and Raf11 are positive regulators of seed dormancy and ABA response. Plant Cell Physiol 56(1):84–97
Lefebvre V, North H, Frey A, Sotta B, Seo M, Okamoto M, Nambara E, Marion-Poll A (2006) Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J 45(3):309–319
Liu S, Sehgal SK, Lin M, Li J, Trick HN, Gill BS, Bai G (2015) Independent mis-splicing mutations in TaPHS1 causing loss of preharvest sprouting (PHS) resistance during wheat domestication. New Phytol 208(3):928–935
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu YG (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot Plants. Mol Plant 8(8):1274–1284
Mao X, Zhang J, Liu W, Yan S, Liu Q, Fu H, Zhao J, Huang W, Dong J, Zhang S, Yang T, Yang W, Liu B, Wang F (2019) The MKKK62-MKK3-MAPK7/14 module negatively regulates seed dormancy in rice. Rice 12(1):2
Matsuoka D, Yasufuku T, Furuya T, Nanmori T (2015) An abscisic acid inducible Arabidopsis MAPKKK, MAPKKK18 regulates leaf senescence via its kinase activity. Plant Mol Biol 87(6):565–575
Miao C, Xiao L, Hua K, Zou C, Zhao Y, Bressan RA, Zhu JK (2018) Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc Natl Acad Sci 115(23):6058–6063
Min MK, Choi EH, Kim JA, Yoon IS, Han S, Lee Y, Lee S, Kim BG (2019) Two clade a phosphatase 2Cs expressed in guard cells physically interact with abscisic acid signaling components to induce stomatal closure in rice. Rice 12(1):37
Mitula F, Tajdel M, Cieśla A, Kasprowicz-Maluśki A, Kulik A, Babula-Skowrońska D, Michalak M, Dobrowolska G, Sadowski J, Ludwików A (2015) Arabidopsis ABA-activated kinase MAPKKK18 is regulated by protein phosphatase 2C ABI1 and the ubiquitin-proteasome pathway. Plant Cell Physiol 56(12):2351–2367
Mukherjee K, Choudhury AR, Gupta B, Gupta S, Sengupta DN (2006) An ABRE-binding factor, OSBZ8, is highly expressed in salt tolerant cultivars than in salt sensitive cultivars of indica rice. BMC Plant Biol 6:18
Nakamura S, Abe F, Kawahigashi H, Nakazono K, Tagiri A, Matsumoto T, Utsugi S, Ogawa T, Handa H, Ishida H, Mori M, Kawaura K, Ogihara Y, Miura H (2011) A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell 23(9):3215–3229
Nakamura S, Pourkheirandish M, Morishige H, Kubo Y, Nakamura M, Ichimura K, Seo S, Kanamori H, Wu J, Ando T, Hensel G, Sameri M, Stein N, Sato K, Matsumoto T, Yano M, Komatsuda T (2016) Mitogen-activated protein kinase kinase 3 regulates seed dormancy in barley. Curr Biol 26(6):775–781
Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, Shinozaki K, Yamaguchi-Shinozaki K (2009) Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 950(7):1345–1363
Pan Y, Chen L, Zhao Y, Guo H, Li J, Rashid MAR, Lu C, Zhou W, Yang X, Liang Y, Wu H, Qing D, Gao L, Dai G, Li D, Deng G (2021a) Natural variation in OsMKK3 contributes to grain size and chalkiness in rice. Front Plant Sci 12:784037
Pan YH, Gao LJ, Liang YT, Zhao Y, Liang HF, Chen WW, Yang XH, Qing DJ, Gao J, Wu H, Huang J, Zhou WY, Huang CC, Dai GX, Deng GF (2021b) OrMKK3 influences morphology and grain size in rice. J Plant Biol 66:1–14
Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF, Cutler SR (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324(5930):1068–1071
Pei ZM, Murata Y, Benning G, Thomine S, Klüsener B, Allen GJ, Grill E, Schroeder JI (2000) Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406(6797):731–734
Peng L, Sun S, Yang B, Zhao J, Li W, Huang Z, Li Z, He Y, Wang Z (2022) Genome-wide association study reveals that the cupin domain protein OsCDP3.10 regulates seed vigour in rice. Plant Biotechnol J 20(3):485–498
Rao KP, Richa T, Kumar K, Raghuram B, Sinha AK (2010) In silico analysis reveals 75 members of mitogen-activated protein kinase kinase kinase gene family in rice. DNA Res 17(3):139–153
Rodriguez MC, Petersen M, Mundy J (2010) Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61:621–649
Sato Y, Takehisa H, Kamatsuki K, Minami H, Namiki N, Ikawa H, Ohyanagi H, Sugimoto K, Antonio BA, Nagamura Y (2013) RiceXPro version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res 41:D1206–D1213
Sethi V, Raghuram B, Sinha AK, Chattopadhyay S (2014) A mitogen-activated protein kinase cascade module, MKK3-MPK6 and MYC2, is involved in blue light-mediated seedling development in Arabidopsis. Plant Cell 26(8):3343–3357
Shu K, Liu XD, Xie Q, He ZH (2016) Two faces of one seed: hormonal regulation of dormancy and germination. Mol Plant 9(1):34–45
Song S, Wang G, Wu H, Fan X, Liang L, Zhao H, Li S, Hu Y, Liu H, Ayaad M, Xing Y (2020) OsMFT2 is involved in the regulation of ABA signaling-mediated seed germination through interacting with OsbZIP23/66/72 in rice. Plant J 103(2):532–546
Sözen C, Schenk ST, Boudsocq M, Chardin C, Almeida-Trapp M, Krapp A, Hirt H, Mithöfer A, Colcombet J (2020) Wounding and insect feeding trigger two independent MAPK pathways with distinct regulation and kinetics. Plant Cell 32(6):1988–2003
Tian X, Wang Z, Li X, Lv T, Liu H, Wang L, Niu H, Bu Q (2015) Characterization and functional analysis of pyrabactin resistance-like abscisic acid receptor family in rice. Rice 8(1):28–28
Torada A, Koike M, Ogawa T, Takenouchi Y, Tadamura K, Wu J, Matsumoto T, Kawaura K, Ogihara Y (2016) A causal gene for seed dormancy on wheat chromosome 4A encodes a MAP kinase kinase. Curr Biol 26(6):782–787
Wang P, Song CP (2008) Guard-cell signalling for hydrogen peroxide and abscisic acid. New Phytol 178(4):703–718
Wang WQ, Xu DY, Sui YP, Ding XH, Song XJ (2022) A multiomic study uncovers a bZIP23-PER1A-mediated detoxification pathway to enhance seed vigor in rice. Proc Natl Acad Sci 119(9):e2026355119
Xi W, Liu C, Hou X, Yu H (2010) MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 22(6):1733–1748
Xing Y, Jia W, Zhang J (2008) AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J 54(3):440–451
Xing Y, Jia W, Zhang J (2009) AtMKK1 and AtMPK6 are involved in abscisic acid and sugar signaling in Arabidopsis seed germination. Plant Mol Biol 70(6):725–736
Yadav SK, Santosh Kumar VV, Verma RK, Yadav P, Saroha A, Wankhede DP, Chaudhary B, Chinnusamy V (2020) Genome-wide identification and characterization of ABA receptor PYL gene family in rice. BMC Genomics 21(1):676
Zang G, Zou H, Zhang Y, Xiang Z, Huang J, Luo L, Wang C, Lei K, Li X, Song D, Din AU, Wang G (2016) The De-Etiolated 1 homolog of Arabidopsis modulates the ABA signaling pathway and ABA biosynthesis in rice. Plant Physiol 171(2):1259–1276
Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, Wang J, Wang H (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7(1):30
Zhou SC et al (2019) OsMKK3, a stress-responsive protein kinase, positively regulates rice resistance to Nilaparvata lugens via phytohormone dynamics. Int J Mol Sci 20(12):3023
Zou M, Guan Y, Ren H, Zhang F, Chen F (2008) A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol 66(6):675–683
Funding
This study was supported by National Natural Science Foundation of China (31971999), Guangdong Basic and Applied Basic Research Foundation (2019A1515011208), Agricultural Competitive Industry Discipline Team Building Project of Guangdong Academy of Agricultural Sciences (202101TD), Guangdong Special Funds for Rural Revitalization (2022-NBH-00-012), Projects of Guangdong Key Laboratory (2020B1212060047), and the Key Field Research and Development Project of Guangdong Province (2022B0202110003).
Author information
Authors and Affiliations
Contributions
XM, CL and QL conceived and designed the experiment. XM and XZ conducted the experiments, performed data analysis, and wrote the manuscript. BS, LJ, JZ, SL, HY, PC, WC, and ZF participated in some parts of the study. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Fig. S1.
The alignment result of target region in OsMKK3/OsMFT2 Crispr lines.
Additional file 2: Fig. S2.
Inhibitory effect of 0.2 μM ABA on mpk7/14. ABA sensitivity index (tissue length under 0.2 μM ABA treatment/length under 0 μM ABA treatment) of ZH11 and mpk7/14 for shoots and roots. Values represent the mean ± SD of three biological replicates (10 germinated seeds for each replicate). The asterisks indicate significant differences compared with ZH11. Student’s t-tests were used to generate p values (*p < 0.05, **p < 0.01).
Additional file 3: Table S1.
Primers used in this study.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mao, X., Zheng, X., Sun, B. et al. MKK3 Cascade Regulates Seed Dormancy Through a Negative Feedback Loop Modulating ABA Signal in Rice. Rice 17, 2 (2024). https://doi.org/10.1186/s12284-023-00679-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-023-00679-4