Abstract
Background
Studies have shown that the effects of maternal nutrition exposure during gestation influence metabolic risk in early life through an epigenetic mechanism. Low glycaemic index (GI) diets benefit both maternal and neonatal gestational outcomes. We hypothesize that maternal dietary GI or glycaemic load (GL) changes during pregnancy impact placental DNA methylation, especially in insulin resistance-related genes.
Methods
From a clinical trial of overweight pregnant women, 12 subjects who successfully reduced their GI and another 12 whose GI increased despite the intervention were selected. A genome-wide differential methylation analysis of placental tissue DNA was conducted, followed by bioinformatic annotation and validation analysis. The distribution of genome-wide differentially methylated regions (DMRs) and CpG sites was described. Six CpG sites in regulatory regions of four insulin-related genes (PLIN1, CPT1B, SSTR4, and CIDEA) were selectively validated by pyrosequencing. Pairwise Spearman correlation analysis was performed to test methylation–phenotype association in an additional 153 subjects from the same trial. Correlation between methylation of significant sites and placental mRNA expression of SSTR4 was also analysed.
Results
Dietary GI decreased by 24.3 (26.2–20.1) in the group who responded appropriately to the intervention and increased by 19.6 (15.2–29.1) in the comparison group. Epigenome-wide analysis identified 108 DMRs and 365 CpG sites with P < 0.05 adjusted by false discovery rate, distributed over all chromosomes. The methylation level of cg05009389 in the 3′ UTR of PLIN1 was negatively correlated with maternal weight gain (ρ = − 0.21, P = 0.027) and increase in insulin levels (ρ = − 0.24, P = 0.015) during gestation. Methylation levels of cg17586860 and cg18197392 in the 5′ UTR region of SSTR4 were negatively correlated with changes in dietary carbohydrate intake (ρ = − 0.24, Ps ≤ 0.006) and GL across gestation (ρ = − 0.23, Ps ≤ .008). This correlation survived the adjustment for maternal factors such as dietary GI, body mass index, and gestational diabetes. Up to 89% of cg18197392 methylation was explained by GL change. Cg14631053 methylation correlated positively with mRNA expression of SSTR4 in the placenta (ρ = 0.20, P = 0.037).
Conclusions
We provide the first evidence that maternal dietary GI changes during gestation may impact placental DNA methylation of insulin regulation genes. This supports the hypothesis that placental methylation may be the epigenetic mechanism through which maternal diet influences the metabolic health of offspring.
Similar content being viewed by others
Background
Maternal obesity or excessive weight gain during pregnancy is recognized as an important issue with a globally increasing trend [1, 2]. Excessive weight gain during pregnancy is associated with multiple maternal and neonatal complications and may have long-term unfavourable effects on the long-term risk of developing metabolic disorders in offspring [3]. A low glycaemic index (GI) diet, as a dietary intervention for pregnant women with high metabolic risk, has been successfully shown to improve maternal gestational outcomes [4, 5], reduce the need for insulin among women with gestational diabetes [6], and reduce neonatal birth weight [7] and incidence of births that are clinically considered “large for gestational age” [4]. Epigenetic processes are suggested to be an important link between maternal gestational dietary exposures and altered offspring metabolism in animal studies [8, 9]. In recent years, emerging evidence from human studies suggests that various types of maternal in utero exposure, such as cigarette smoking [10,11,12], gestational diabetes [13, 14], insufficient maternal weight gain [15], and even maternal anxiety [16], can influence methylation in the placental tissue or cord blood. Some gene methylation changes in these tissues related to maternal gestational diabetes are also linked with adiposity-related outcomes in childhood [17]. A recent study reported that maternal glycaemia during pregnancy is correlated with placental DNA methylation levels of brown adipose tissue-related genes, and variations in DNA methylation levels of these same genes are correlated with cord blood leptin levels [18]. Another study reported that higher retinoid X receptor alpha (RXRA) gene promoter methylation levels in the umbilical cord tissue were associated with lower maternal carbohydrate intake in early pregnancy and child adiposity at 9 years of age [19]. However, evidence is lacking as to whether maternal GI diet changes during gestation are associated with offspring gene methylation changes.
Results
Clinical characteristics of the two groups of study samples for genome-wide analysis and validation analysis are shown in Table 1. In terms of the discovery study sample for genome-wide differential methylation analysis, the GI decreased from 67.6 to 42.8 with a change of 24.3 (26.2–20.1) in the decreased GI group and increased from 51.2 to 73.5 with a change of 19.6 (15.2–29.1) in the increased GI group across the gestational period. There were no additional significant differences in the other characteristics at baseline or in pregnancy outcomes at delivery between the two groups. In the validation study sample, the GI decreased 1.1 on average but with larger variability. The GI increased in 76.0% of subjects and decreased in 24.0% of subjects (data were not shown).
The genome-wide differential methylation analysis (step 1) identified 108 DMRs with adjusted P < 0.05 (raw DMR P ranged from 10−24 to 10−3), including 365 CpG sites, and mapped to 95 genes (details in Additional file 1). These CpG sites were distributed across all autosomal chromosomes, with all possible relationships to CpG islands, including within CpG islands, the open sea, the shelf, or the shore of CpG islands (Fig. 1). Among these sites, 149 (40.8%) sites were hypomethylated and 216 (59.2%) sites were hypermethylated. After FDR adjustment, 18 enriched GO terms in the interaction network were genomically significant (FDR < 0.05) (Additional file 2) (step 2). The most significant terms referred to biological processes, such as positive regulation of signalling.
From 365 differentially methylated sites, those in regulatory regions of insulin regulation genes indicated by GO term analysis were selected for validation analysis (step 3). Ten CpG sites in PLIN1, CPT1B, SSTR4, and CIDEA that were validated were hypomethylated in the decreased GI group (Table 2). Among which, four sites failed for technical reasons and the remaining six sites (all located in CpG islands) were examined by pyrosequencing in the validation cohort and were included in further methylation–phenotype correlation analysis, one for PLIN1, CPT1B, and CIDEA and three for SSTR4. The pairwise methylation–phenotype analyses revealed several weak correlations (Table 3): cg05009389 in the 3′ UTR of PLIN1 was negatively correlated with maternal gestational weight gain (ρ = − 0.21, P = 0.027) and an increase of insulin across gestation (ρ = − 0.24, P = 0.015), cg17586860 and cg18197392 in the 5′ UTR region of SSTR4 (transcription starting site, TSS200) were negatively correlated with the change in carbohydrate intake (ρ = − 0.24, Ps ≥ 0.006), and GL changes across the gestation period (ρ = − 0.23, Ps ≤ 0.008). We did not find significant correlations of cg14631057 methylation with any clinical phenotypes; however, it showed moderate correlations with the methylation of the other two sites in the same CpG island in the promoter of SSTR4 (ρ was 0.53–0.59, Ps < 0.0001). As shown in Table 4, further multivariate regression analyses showed that the GI at the first interview and GI change during gestation, as well as BMI and gestational diabetes mellitus (GDM), explained 9.3% of the total variance of cg17586860 methylation. The GI change was independently associated with cg17586860 methylation levels (β = − 0.36, P = 0.003), explaining 72.8% of the variance after adjustment for covariates. No significant results were found for the other CpG sites (Table 4).
Among the three CpG sites in the CpG island of the SSTR4 promoter, the cg14631053 methylation levels were correlated with the placental mRNA expression of SSTR4 (ρ = 0.20, P = 0.037), and no significant correlations were found for the other two CpG sites.
Discussion
By using genome-wide DNA differential methylation analyses followed by validation analyses, we provide evidence suggesting that maternal dietary glycaemic changes during gestation are associated with placental DNA methylation changes in insulin resistance-related genes. We identified 108 DMRs across the whole genome including 365 CpG sites and 95 genes. By using a larger sample size, we also found consistent weak correlations between methylation levels of CpG sites in the promoter region of four insulin resistance-related genes with maternal dietary glycaemic change and with changes of some other related clinical phenotypes across gestation: methylation of one CpG site in the 3′ UTR of PLIN1 gene and two sites of a CpG island close to TSS of SSTR4 gene are associated with maternal changes of dietary GI, GL, weight gain, and insulin levels during gestation. In addition, methylation of one CpG site from the same CpG island in SSTR4 is weakly correlated with the placental mRNA expression of SSTR4 gene. These results suggest that placental DNA methylation may be altered as a response to significant changes in maternal diet GI, even in a short period of gestation (approximately 20 weeks). The methylation and gene expression alterations in regulatory regions of insulin resistance-related genes in the placental tissue may be the link between maternal diet modifications with foetal outcomes or future metabolic risks, which is consistent with some previous clinical studies.
One of our findings is that maternal dietary glycaemic changes are associated with methylation alterations in hundreds of genes across the genome. In combination with previous studies, these findings support the epigenetic impact of maternal nutritional exposure during gestation on offspring metabolic risk. Some previous studies focused on the impact of maternal dietary protein and fat intake [8, 9, 20]. Godfrey et al. [21] reported associations of lower maternal carbohydrate intake in early pregnancy and hypermethylated RXRA genes in the umbilical cord tissue of healthy neonates and the association between this hypermethylation with children’s fat mass at age 9. In the current study, based on the placental tissue instead of the umbilical cord tissue, we did not find significantly differential methylation of the RXRA gene between pregnant women with distinct and opposite dietary glycaemic changes. Ruchat et al. reported that maternal GDM epigenetically affects genes predominantly involved in metabolic diseases; however, the placental tissue and cord blood share only 25% of differentially methylated CpG sites [22]. In our study, PLIN1 and SSTR4, whose methylation patterns are associated with maternal dietary GI change and related phenotype change, are not in the gene list of the previous study [22]. Park et al. found that the obesity status of women influences gene methylation changes in response to folic acid supplementation [23]. Similarly, only some of the genes had similar methylation changes in the cord blood sample and placental samples. It is known that DNA methylation is widely dynamic and tissue-specific during the development of humans [22], the cellular composition of placentas varies among individuals [3], and the methylation profiles differ in different cell types in human placentas [24]. Although the collection of the placental tissue followed standardized procedure, the heterogeneity of sampling by different investigators may have led to heterogeneity in the cell type composition of the placental tissue and may have an impact on methylation levels of selected genes. However, this impact on methylation is likely to be non-directional, making it less likely to be a source of bias to the weak associations found in this study.
The SSTR4 gene encodes the type 4 receptor of somatostatin that exerts inhibitory effects on all endocrine and exocrine secretions in humans, including its role as an endogenous inhibitor of cell proliferation [15] and function in certain areas of the central nervous system, such as motor, sensory, behavioural, cognitive, and autonomic effects [20]. The SSTR4 gene is expressed in human placental tissue [21]. The CpG site, cg17586860, survived the two-stage association analysis and is correlated with maternal GL change and with methylation patterns of other sites (cg14631053 and cg18197392). This site is located in the TSS200 region of SSTR4, where sequences are with potentially important functions, such as binding sites of several transcription factors and elements required for tissue-specific promoter activity [25, 26]. Previous studies conducted transfection experiments focusing on this region but did not indicate any effects of possible regulators on transcriptional regulation of SSTR4. Sequence analysis of the SSTR4 gene did not find the proximal 5′ UTR to contain any potential TATA or CAAT boxes but that it was highly GC-rich within the first 300 bps [26], which contains a CpG island. The correlation we find that the reduced methylation of cg17586860 in this island in relation to greater dietary GL decrease (weak negative correlation) may support the hypothesis that maternal dietary glycaemic change may have a favourable impact on SSTR4 mRNA expression through the alteration of methylation status of promoter region and possible further effects on foetal development. Unfortunately, placental SSTR4 protein expression was not examined in the current study. It still remains unclear how the gene methylation and mRNA expression modifications observed may affect the structure or the function of the placenta or other potential target organs since the evaluation of the proteins regulated by the genes studied is not available. Future study is expected to examine the effects of reduced expression of SSTR4, possibly including the enhancement of various endocrine and exocrine secretions in the placenta.
Another interesting finding is the differential methylation of cg05009389 in the 3′ UTR of the placenta PLIN1 gene in relation with extreme maternal dietary GI change during pregnancy. We found weak correlations between reduced methylation with greater maternal insulin increase and body weight gain. The PLIN1 gene encodes perilipin-1, which coats lipid storage droplets in adipocytes and functions as inhibitors of lipolysis [20]. Godfrey et al. indicated a gene–nutrient interaction between a PLIN1 polymorphism and dietary intake of saturated fat and carbohydrates and their effect on insulin resistance levels [21]. Loss-of-function mutations in PLIN1 have been reported to lead to familial partial lipodystrophy, severe insulin resistance, and diabetes [22]. PLIN1 promoter methylation was observed to be inversely correlated with PLIN1 mRNA expression and lipolytic activity [23]. This evidence indicates epigenetic regulation of the gene and its function. The findings of our study may support a possible epigenetic mechanism of the influence of maternal dietary glycaemic change on offspring’s future risk of lipid metabolic disorders. Unfortunately, PLIN1 is commonly expressed in adipose tissue and not in human placenta tissue, so examining its expression was not possible in the current study.
Strengths of our study include, firstly, the use of human placental tissue to assess the epigenetic effects of maternal dietary GI change during pregnancy on DNA methylation in offspring. Secondly, the application of the Infinium 450K BeadChip technology to assess genome-wide methylation profiles offered greatly improved genomic coverage over the earlier 27K platform. Thirdly, in the current study, the maternal dietary glycaemic change was as a result from counselling, not from directly receiving low glycaemic food. Its associations with placental tissue gene methylation support the importance of maternal lifestyle modification during gestation.
This study has several limitations. Firstly, the subjects included in our study were from a randomized controlled trial and limited as to whose placental tissues were successfully collected and processed. Thus, the interpretation of associations identified in the current study sample should be taken with caution. Secondly, the sample size for KEGG pathway and GO analyses was relatively small. Thirdly, only the subgroup of CpG sites (in the regulatory region of genes that related to insulin resistance) was chosen from the genome-wide differential methylation analysis for methylation–phenotype correlation analyses in the larger study sample.
Conclusions
We report for the first time that maternal dietary GI changes are associated with offspring’s placental DNA methylome, including in insulin resistance-related genes. Methylation in the promoter regions of PLIN1 and SSTR4 is correlated with maternal changes in dietary GI, GL, and other clinical phenotypes. This evidence supports the idea that placenta methylation alteration may be the epigenetic mechanism underlying favourable effects of maternal dietary habit changes on their offspring. Future work is needed to determine the relevance of epigenetic changes in human placenta tissue to the change of maternal GI during pregnancy.
Methods
Participants
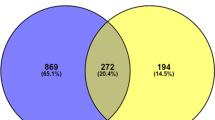
Participants were selected from a lifestyle intervention study (clinicaltrials.gov, NCT01628835), which was carried out at two maternity and child care centres in southeast China from June 2012 to October 2015. In brief, overweight pregnant women were recruited at the first antenatal examination and received dietary counselling three times during gestation; the control group received standard dietary counselling according to the nationwide nutritional recommendations, and the experimental group additionally received an accurate assessment of their personal dietary glycaemic index and advice about low glycaemic diets, when needed. Overall, all mother–child pairs with completed data in terms of prenatal exposure (including their individual change in dietary GI on each visit) and outcomes at birth (including a collection of placental tissue samples) were included in the current study (n = 177). Of these, the 12 subjects whose GI decreased the most remarkably during gestation were chosen as the “GI decrease group,” and 12 subjects whose GI increased most remarkably, despite intervention to lower their GI, were chosen as the “GI increase group,” for genome-wide differential methylation analysis. The remaining 153 mother–child pairs were treated as a validation sample for methylation-phenotype association analysis (Fig. 2).
Maternal dietary GI measurements during gestation
Maternal dietary data were collected by 24-h dietary recall at three face-to-face dietary consultation interviews. These interviews were incorporated as a part of the subjects’ routine antenatal care visits to improve compliance. The baseline interview was conducted at the first antenatal visit. The second interview was arranged to coincide with the conventional 75-g oral glucose tolerance test (OGTT) during the 24th to 28th week, and the third interview coincided with the usual liver function test during the 34th to 36th week. Dietary GL, GI, and nutrients were evaluated using a customized Excel worksheet that incorporated the Chinese [27] and international food composition tables and published GI values [28,29,30]. The change in GI during pregnancy was calculated as the GI at the third interview minus the GI at the first interview.
Data and biological sample collection
Demographic, clinical, and anthropometric data were collected on the first visit (baseline). Body mass index (BMI) was calculated as weight (kg)/[height (m)]2 and categorized according to Chinese categories (underweight < 18.5 kg/m2, normal weight 18.5–23.9 kg/m2, overweight 24.0–27.9 kg/m2, and obese ≥ 28 kg/m2) [31]. The gestational age was estimated from the last self-reported menstrual period and corrected by the first routine ultrasound examinations around the 16th week of gestation. Routine lab examination data, including plasma glucose, serum insulin, and glycosylated haemoglobin, were obtained from the hospital information system. The insulin resistance index was calculated (under a model of homeostasis) using the following formula [32]: [fasting plasma glucose (mmol/L)] × [fasting insulin (mIU/L)]/22.5. GDM was defined as having one or more abnormal values from the 2-h 75-g OGTT between 24 and 28 weeks of gestation, with cutoff values of 5.1 mmol/L for fasting, 10.0 mmol/L for 1 h, and 8.5 mmol/L for 2 h [33]. Gestational weight gain was calculated as the body weight at delivery minus the body weight at baseline.
Placentas were collected immediately after delivery by midwives from the medical institution according to standard protocols. Briefly, biopsies of six points of the inner region of the placenta around the umbilical cord (the circumference of the radius was 4 cm) were obtained, rinsed by normal saline, immersed into a tube with RNAlater™ stabilization solution, and preserved at − 20 °C. Within a week, the samples were moved to a − 80 °C freezer.
Genome-wide DNA differential methylation analysis and validation
Firstly, the genome-wide differential methylation analysis was used to identify significant DMRs, CpG sites, and genes (step 1). Secondly, we performed GO term enrichment analysis to identify CpG sites in genes that are related to insulin regulation (step 2). Thirdly, we selected CpG sites for validation and correlation analysis with maternal phenotypes in the regulatory regions of the selected insulin-related genes since these CpG sites are more likely to be functional to impact gene expression (step 3).
Genomic DNA was isolated from ground-up placentas using the QIAamp DNA Mini Kit (QIAGEN, Germany). Bisulfite conversion using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA) was carried out according to the manufacturer’s protocols. Genome-wide DNA methylation levels of 24 subjects were examined by Infinium 450K Human Methylation BeadChip (Illumina, San Diego, CA, USA), following the standard manufacturer’s manual. DNA from the two groups of subjects were randomized and assigned to two chips (12 samples on each chip) in a 1:1 ratio and processed by the same technician at the same time to minimize batch and chip effects of the two compared groups. On each chip, 12 DNA samples were randomly assigned to cells.
Signals of all 485,577 CpGs were analysed to distinguish from background with detection P values less than 0.01, according to the MinifiR package [34], which also estimated the methylation β value which is interpreted as the proportion of DNA molecules in the sample with methylation at a given site and ranged between 0 (unmethylated) and 1 (fully methylated). Probes with a bead count less than 3 in 5% or more of the samples, probes on chromosome X or Y, and the 65 control probes were also excluded. Finally, 434,958 CpG sites were available for differential analysis. The beta mixture quantile dilation (BMIQ) method was used for normalization [35]. Probe Lasso [36] using the ChAMP R package (version 1.8.2, http://bioconductor.org/packages/ChAMP/) was employed to identify differentially methylated regions (DMRs), and FDR-adjusted DMR P values were determined using a method by Benjamini and Hochberg [37]. Gene oncology (GO) terms and protein interaction (KEGG) enrichment analyses were performed for all CpG sites involved in significant DMRs (adjusted P < 0.05).
From 365 differentially methylated sites indicated by stage 1 analysis, those in the regulatory regions (promoter, 5′ UTR, and 3′ UTR) of insulin regulation genes and indicated by GO term analysis were selected for validation analysis. Literature research was undertaken to check the functions of these genes. Ten CpG sites in the promoter region or in the 3′ UTR region of four insulin resistance-related genes were selected for further examination and methylation–phenotype association analysis. For quality control, CpG sites in the regulatory region (3′ UTR or 5′ UTR) of the selected genes were validated using pyrosequencing in the discovery cohort (n = 24 subjects) followed by further analysis using all subjects with available placenta tissue samples (n = 153 subjects).
mRNA expression analysis of the SSTR4 gene
Total RNA was isolated from 50 to 100 mg placental tissue (stored at − 80 °C) using the Qiagen RNA mini kit, according to the manual’s instructions. Complementary DNA (cDNA) was synthesized using TAKARA reverse transcription kits (RR036A Takara PrimeScript™ RT Master Mix, Japan). The primers used were designed using Primer Premier 5.0 software. Primer sequences were as follows: SSTR4 (forward) 5′-CCTTCGCTACGCCAAGATGA-3′ and (reverse) 5′-GAGACAGAAGACGCTGGTGAA-3′, yielding a PCR product length of 196 bp. GAPDH primer was (forward) 5′-GAAGGTCGGAGTCAACGGATT-3′ and (reverse) 5′-TGCTGATGATCTTGAGGCTGTT-3′, yielding a PCR product length of 434 bp. Total RNA mixed with RNase-free water (8 μL) and 2 μL PrimeScript RT Master Mix was added to a tube placed in an ice bath, and the contents settled to the bottom of the tube by spinning briefly for 3–5 s. The tube was placed in a 37 °C water bath (ABI PCR 2720) for 15 min and an 85 °C water bath for 5 s and then cooled on ice. cDNA was stored at − 20 °C until use.
RT-PCR was performed in triplicate to determine the SSTR4 mRNA expression of the placental tissue in 153 subjects using the LightCycler 480 II (Roche). For amplification by PCR, 6 μL RNase-free water, 10 μL real-time PCR mix, 2 μL cDNA, 1 μL forward primer (10 μM), and reverse primer (10 μM) were added. Using GAPDH as an internal control, the amplification of the SSTR4 gene was performed in the same tube under the following thermal cycling conditions: initial denaturation at 95 °C for 1 min, followed by 45 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s and extension at 72 °C for 30 s, and the last extension at 72 °C for 30 s. The melting curve included 1 cycle at 95 °C for 5 s and 65 °C for 1 min. The expression of SSTR4 mRNA was estimated by the 2-ΔΔCT method using G038 as controls.
Placenta DNA methylation–maternal phenotype association analysis
Continuous phenotypes are described by their mean ± SD or median (interquartile range) where necessary, and categorical data are described by percentages. Comparisons of the characteristics of pregnant women between the control and the decreased GI group were performed by using independent sample t tests for continuous variables and chi-squared tests for categorical variables. Correlations between methylation levels, gene expression levels, and maternal phenotypes were analysed using a pairwise Spearman correlation analysis. Maternal phenotypes included changes of dietary GI, GL, fibre, carbohydrates, and serum insulin, as well as body weight gain across gestation. Contributions of the GI diet change to CpG site methylation levels were evaluated by explained R2 of stepwise multiple regression to adjust possible covariates. P < 0.05 was chosen as the significant level.
The protocol of the study was approved by the institutional ethics committee. Written informed consent forms were obtained from each subject at recruitment before any data collection.
Abbreviations
- BMI:
-
Body mass index
- BMIQ:
-
Beta mixture quantile dilation
- DMR:
-
Differentially methylated region
- GDM:
-
Gestational diabetes mellitus
- GI:
-
Glycaemic index
- GL:
-
Glycaemic load
- GO:
-
Gene ontology
- OGTT:
-
Oral glucose tolerance test
- TSS:
-
Transcription starting site
References
Hyperglycemia and Adverse Pregnancy Outcome (hapo) study: associations with neonatal anthropometrics. Diabetes. 2009;58:453–9.
Fraser A, Tilling K, Macdonald-Wallis C, Sattar N, Brion MJ, Benfield L, Ness A, Deanfield J, Hingorani A, Nelson SM, Smith GD, Lawlor DA. Association of maternal weight gain in pregnancy with offspring obesity and metabolic and vascular traits in childhood. Circulation. 2010;121:2557–64.
Mayhew TM, Charnock-Jones DS, Kaufmann P. Aspects of human fetoplacental vasculogenesis and angiogenesis. III. Changes in complicated pregnancies. Placenta. 2004;25:127–39.
Moses RG, Luebcke M, Davis WS, Coleman KJ, Tapsell LC, Petocz P, Brand-Miller JC. Effect of a low-glycaemic-index diet during pregnancy on obstetric outcomes. Am J Clin Nutr. 2006;84:807–12.
Moses RG, Luebke M, Petocz P, Brand-Miller JC. Maternal diet and infant size 2 y after the completion of a study of a low-glycaemic-index diet in pregnancy. Am J Clin Nutr. 2007;86:1806.
Moses RG, Barker M, Winter M, Petocz P, Brand-Miller JC. Can a low-glycaemic index diet reduce the need for insulin in gestational diabetes mellitus? A randomized trial. Diabetes Care. 2009;32:996–1000.
Scholl TO, Chen X, Khoo CS, Lenders C. The dietary glycaemic index during pregnancy: influence on infant birth weight, fetal growth, and biomarkers of carbohydrate metabolism. Am J Epidemiol. 2004;159:467–74.
Burdge GC, Slater-Jefferies J, Torrens C, Phillips ES, Hanson MA, Lillycrop KA. Dietary protein restriction of pregnant rats in the f0 generation induces altered methylation of hepatic gene promoters in the adult male offspring in the f1 and f2 generations. Br J Nutr. 2007;97:435–9.
Dunn GA, Bale TL. Maternal high-fat diet effects on third-generation female body size via the paternal lineage. Endocrinology. 2011;152:2228–36.
Nicole W. Pattern of clues: evidence of distinct DNA methylation in newborns of smoking women. Environ Health Perspect. 2012;120:a402.
Suter MA, Aagaard K. What changes in DNA methylation take place in individuals exposed to maternal smoking in utero? Epigenomics. 2012;4:115–8.
Joubert BR, Haberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, Huang Z, Hoyo C, Midttun O, Cupul-Uicab LA, Ueland PM, Wu MC, Nystad W, Bell DA, Peddada SD, London SJ. 450k epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect. 2012;120:1425–31.
Finer S, Mathews C, Lowe R, Smart M, Hillman S, Foo L, Sinha A, Williams D, Rakyan VK, Hitman GA. Maternal gestational diabetes is associated with genome-wide DNA methylation variation in placenta and cord blood of exposed offspring. Hum Mol Genet. 2015;24:3021–9.
Nomura Y, Lambertini L, Rialdi A, Lee M, Mystal EY, Grabie M, Manaster I, Huynh N, Finik J, Davey M, Davey K, Ly J, Stone J, Loudon H, Eglinton G, Hurd Y, Newcorn JH, Chen J. Global methylation in the placenta and umbilical cord blood from pregnancies with maternal gestational diabetes, preeclampsia, and obesity. Reprod Sci. 2014;21:131–7.
Reubi JC, Laissue JA. Multiple actions of somatostatin in neoplastic disease. Trends Pharmacol Sci. 1995;16:110–5.
Vangeel EB, Pishva E, Hompes T, van den Hove D, Lambrechts D, Allegaert K, Freson K, Izzi B, Claes S. Newborn genome-wide DNA methylation in association with pregnancy anxiety reveals a potential role for gabbr1. Clin Epigenetics. 2017;9:107.
Yang IV, Zhang W, Davidson EJ, Fingerlin TE, Kechris K, Dabelea D. Epigenetic marks of in utero exposure to gestational diabetes and childhood adiposity outcomes: the EPOCH study. Diabet Med. 2018;35:612–20.
Cote S, Gagne-Ouellet V, Guay SP, Allard C, Houde AA, Perron P, Baillargeon JP, Gaudet D, Guerin R, Brisson D, Hivert MF, Bouchard L. Ppargc1alpha gene DNA methylation variations in human placenta mediate the link between maternal hyperglycemia and leptin levels in newborns. Clin Epigenetics. 2016;8:72.
Lillycrop K, Murray R, Cheong C, Teh AL, Clarke-Harris R, Barton S, Costello P, Garratt E, Cook E, Titcombe P, Shunmuganathan B, Liew SJ, Chua YC, Lin X, Wu Y, Burdge GC, Cooper C, Inskip HM, Karnani N, Hopkins JC, Childs CE, Chavez CP, Calder PC, Yap F, Lee YS, Chong YS, Melton PE, Beilin L, Huang RC, Gluckman PD, Harvey N, Hanson MA, Holbrook JD, Godfrey KM. Anril promoter DNA methylation: a perinatal marker for later adiposity. EBioMedicine. 2017;19:60–72.
Lillycrop KA, Phillips ES, Torrens C, Hanson MA, Jackson AA, Burdge GC. Feeding pregnant rats a protein-restricted diet persistently alters the methylation of specific cytosines in the hepatic ppar alpha promoter of the offspring. Br J Nutr. 2008;100:278–82.
Godfrey KM, Sheppard A, Gluckman PD, Lillycrop KA, Burdge GC, McLean C, Rodford J, Slater-Jefferies JL, Garratt E, Crozier SR, Emerald BS, Gale CR, Inskip HM, Cooper C, Hanson MA. Epigenetic gene promoter methylation at birth is associated with child’s later adiposity. Diabetes. 2011;60:1528–34.
Liang P, Song F, Ghosh S, Morien E, Qin M, Mahmood S, Fujiwara K, Igarashi J, Nagase H, Held WA. Genome-wide survey reveals dynamic widespread tissue-specific changes in DNA methylation during development. BMC Genomics. 2011;12:231.
Ruchat SM, Houde AA, Voisin G, St-Pierre J, Perron P, Baillargeon JP, Gaudet D, Hivert MF, Brisson D, Bouchard L. Gestational diabetes mellitus epigenetically affects genes predominantly involved in metabolic diseases. Epigenetics. 2013;8:935–43.
Bianco-Miotto T, Mayne BT, Buckberry S, Breen J, Rodriguez Lopez CM, Roberts CT. Recent progress towards understanding the role of DNA methylation in human placental development. Reproduction. 2016;152:R23–30.
Petersenn S. Structure and regulation of the growth hormone secretagogue receptor. Minerva Endocrinol. 2002;27:243–56.
Petersenn S, Rasch AC, Presch S, Beil FU, Schulte HM. Characterization of the human somatostatin receptor type 4 promoter. Mol Cell Endocrinol. 2002;188:75–83.
Yang Y, Wang G, Pei X. Chinese table of food composition. Beijing: Peking university; 2002.
Foster-Powell K, Holt SH, Brand-Miller JC. International table of glycaemic index and glycaemic load values: 2002. Am J Clin Nutr. 2002;76:5–56.
Neuhouser ML, Tinker LF, Thomson C, Caan B, Horn LV, Snetselaar L, Parker LM, Patterson RE, Robinson-O'Brien R, Beresford SA, Shikany JM. Development of a glycaemic index database for food frequency questionnaires used in epidemiologic studies. J Nutr. 2006;136:1604–9.
T.C R, I, Seelig M. Comparable data on food and nutrient intake and physical measurements from the 1983, 1985 and 1995 national nutrition surveys. Australian food and nutrition monitoring unit. 2001.
Zhou B, Coorperative Meta-analysis Group of China Obesity Task Force. Predictive value of body mass index and waist circumferene to risk factors of related diseases in Chinese adult population. Zhonghua Liu Xing Bing Xue Za Zhi. 2002;23:5–10.
Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–9.
Puri V, Ranjit S, Konda S, Nicoloro SM, Straubhaar J, Chawla A, Chouinard M, Lin C, Burkart A, Corvera S, Perugini RA, Czech MP. Cidea is associated with lipid droplets and insulin sensitivity in humans. Proc Natl Acad Sci U S A. 2008;105:7833–8.
Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA. Minfi: a flexible and comprehensive bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–9.
Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, Beck S. A beta-mixture quantile normalization method for correcting probe design bias in illumina infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–96.
Butcher LM, Beck S. Probe lasso: a novel method to rope in differentially methylated regions with 450k DNA methylation data. Methods. 2015;72:21–8.
Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–8.
Acknowledgements
The authors thank the nurses and technicians who are from the Kunshan Maternity and Child Care Center and the International Peace Maternity & Child Health Hospital of China Welfare Institute, not included in the author list for collecting the data and samples. We also thank all participants. We thank Professor Joe Terwilliger from Columbia University Medical Center for the help in language editing.
Funding
This work was financially supported by the National Science Foundation of China (Grant No. 81273168) and the National Key Scientific and Development Projects (Grant No. 2016YFC1000506).
Availability of data and materials
The datasets used and analysed in this study are available from the corresponding author upon request.
Author information
Authors and Affiliations
Contributions
WY was the principal investigator of the study; contributed to the conception, study design, draft, and critical approval of the draft; and gave input at all stages of the study. WY, YZ, and ND performed the data analyses and made the statistical charts. LPW, WY, CL, LLW, PG, YX, JY, YS, and QZ participated in the field work of the two centres and assisted with the acquisition of routine clinical data. QZ, KM, DN, and YZ contributed to the acquisition and management of data. All authors approved the final manuscript to be submitted.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study has been approved by the Institutional Ethics Committee, and written informed consent was obtained from all subjects (approval number 2014-071).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Significant DMRs and CpG sites based on the genome-wide differential methylation analysis (FDR-adjusted P < 0.05) and annotation information. (XLSX 148 kb)
Additional file 2:
Significant GO terms by enrichment analysis based on CpG sites from four selective genes related to insulin regulation. (XLS 4 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yan, W., Zhang, Y., Wang, L. et al. Maternal dietary glycaemic change during gestation influences insulin-related gene methylation in the placental tissue: a genome-wide methylation analysis. Genes Nutr 14, 17 (2019). https://doi.org/10.1186/s12263-019-0634-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12263-019-0634-x