Abstract
Background
Diet has a great impact on the risk of developing features of metabolic syndrome (MetS), type 2 diabetes mellitus (T2DM), and cardiovascular diseases (CVD). We evaluated whether a long-term healthy Nordic diet (ND) can modify the expression of inflammation and lipid metabolism-related genes in peripheral blood mononuclear cells (PBMCs) during a 2-h oral glucose tolerance test (OGTT) in individuals with MetS.
Methods
A Nordic multicenter randomized dietary study included subjects (n = 213) with MetS, randomized to a ND group or a control diet (CD) group applying an isocaloric study protocol. In this sub-study, we included subjects (n = 89) from three Nordic centers: Kuopio (n = 26), Lund (n = 30), and Oulu (n = 33) with a maximum weight change of ±4 kg, high-sensitivity C-reactive protein concentration ≤10 mg L−1, and baseline body mass index <39 kg m−2. PBMCs were isolated, and the mRNA gene expression analysis was measured by quantitative real-time polymerase chain reaction (qPCR). We analyzed the mRNA expression changes of 44 genes before and after a 2hOGTT at the beginning and the end of the intervention.
Results
The healthy ND significantly down-regulated the expression of toll-like receptor 4 (TLR4), interleukin 18 (IL18), and thrombospondin receptor (CD36) mRNA transcripts and significantly up-regulated the expression of peroxisome proliferator-activated receptor delta (PPARD) mRNA transcript after the 2hOGTT compared to the CD.
Conclusions
A healthy ND is able to modify the gene expression in PBMCs after a 2hOGTT. However, more studies are needed to clarify the biological and clinical relevance of these findings.
Similar content being viewed by others
Background
The metabolic syndrome (MetS) is a cluster of risk factors increasing the risk of type 2 diabetes mellitus (T2DM) and cardiovascular diseases (CVD) (Alberti et al. 2009). Obesity, insulin resistance (IR), and T2DM are associated with chronic low-grade inflammation (Bastard et al. 2006; Wellen and Hotamisligil 2005), which plays a pivotal role in all phases of atherosclerosis (Libby et al. 2009). Diet has a great impact on the risk of MetS, T2DM, and CVD (Alberti et al. 2009; Mozaffarian et al. 2011). Thus, it is crucial to understand the role of diet and dietary compounds on inflammation in the development of these diseases.
A healthy Nordic diet (ND) has been shown to improve lipid profile among hyper-cholesterolemic subjects (Adamsson et al. 2011). The Systems Biology in Controlled Dietary Interventions and Cohort Studies (SYSDIET) study was a multicenter randomized dietary study in individuals with features of MetS. A healthy ND with whole-grain products, berries, fruits and vegetables, rapeseed oil, three fish meals per week, and low-fat dairy products was compared to an average Nordic diet served as control diet (CD) (Uusitupa et al. 2013). In the SYSDIET study, we showed that an isocaloric healthy ND improved the lipid profile, low-grade inflammation, and ambulatory blood pressure among subjects with MetS (Uusitupa et al. 2013; Brader et al. 2014). No changes in glucose metabolism were observed since it may be difficult to improve glucose metabolism in established MetS without attendant weight loss and very distinct changes in the diet (Uusitupa et al. 2013).
The peripheral blood mononuclear cells (PBMCs) include monocytes and lymphocytes, which are cells central in inflammation. These cells circulate in the body and are exposed to nutrients, bioactive food components, and metabolic tissues. Alterations in gene expression levels in these cells may therefore reflect systemic health (Afman et al. 2014). It has been shown that long-term dietary intervention studies change the gene expression of inflammatory genes and genes involved in lipid metabolism (Myhrstad et al. 2014; van Dijk et al. 2012a; De Mello et al. 2009; Bouwens et al. 2009), suggesting that PMBCs are a good model system identifying early risk markers (Visvikis-Siest et al. 2007) and are sensitive to dietary changes.
Stress responses can be more informative than static homeostasis on nutrition-related health. An acute glucose load of a 2-h oral glucose tolerance test (OGTT) is such a stress response, monitors the ability of the body to respond to glucose intake, and is primarily used for addressing the degree of glucose tolerance and insulin resistance (van Ommen et al. 2009). Several studies have shown that an OGTT (Choi et al. 2012; Kempf et al. 2007; Aljada et al. 2006) as well as fat challenge tests (Bouwens et al. 2010; Cruz-Teno et al. 2012; van Dijk et al. 2012b; Myhrstad et al. 2011) modulate the gene expression of inflammatory genes in leucocytes and mononuclear cells. PBMCs also reflect the immune component of the white adipose tissue transcriptome after OGTT and after oral lipid tolerance test (O’Grada et al. 2014), and thus, changes in PBMC gene expression may act as biomarkers of metabolic health not only in the fasting state but also in the postprandial state (O’Grada et al. 2014).
Changes in glucose and lipid homeostasis by acute challenge tests are linked to inflammation. It has been shown in dietary intervention studies that the quality of diet affects OGTT response and improves insulin sensitivity and glucose tolerance in individuals with MetS (Laaksonen et al. 2005; Paniagua et al. 2007). However, no previous study has examined the long-term effect of a dietary intervention on the OGTT response using PBMCs and gene expression analysis.
The main aim of this sub-population of the SYSDIET study was to investigate if a long-term (18–24 weeks) healthy ND could modify the expression of inflammation and lipid metabolism-related genes in PBMCs during 2hOGTT in individuals with MetS.
Results
Characteristics of the subjects
At baseline, no differences were observed between the CD and ND groups related to age, BMI, serum lipids, glucose, insulin, circulating inflammation markers, lipid-lowering drugs, antihypertensive drugs, smoking, and MetS (Table 1). The change in glucose, insulin, triglycerides, and free fatty acids from 0h (fasting) to 2h (after OGTT) was not significantly different between the CD and ND groups (P = 0.330, P = 0.845, P = 0.196, and P = 0.681, respectively) (Fig. 1).
Changes in glucose, insulin, triglycerides, and free fatty acids from 0h to 2hOGTT after intervention. The effect of the healthy ND compared to the CD on changes in glucose (a), insulin (b), triglycerides (c), and free fatty acids (d) from 0 h to 2hOGTT after intervention. The effect of the independent variable study group is adjusted for changes in glucose, insulin, triglycerides, or free fatty acids at baseline, study centers, gender, age (log10 transformed), and body weight at the end of the study. Box plots show the medians with 25th and 75th percentiles. Whiskers express the 1.5 × interquartile range
Dietary data
The dietary intake of this sub-population is shown in Table 2. The results are in line with the original analysis with the whole SYSDIET study population (Uusitupa et al. 2013). The intake of polyunsaturated fatty acids was higher and of saturated fatty acids lower in the ND compared to the CD group. Further, α-linolenic acid, fiber, β-carotene, vitamin C, vitamin E, folate, potassium, and magnesium intake were higher in the ND versus the CD group.
Changes in 2hOGTT gene expression response at baseline
Since it is well known that glucose uptake inhibits pyruvate dehydrogenase kinase, isozyme 4 (PDK4), PDK4 mRNA expression in PBMCs was used as a positive control for the 2hOGTT response. A significant down-regulation of PDK4 mRNA expression from 0h (fasting) to 2h (after OGTT) was observed in the whole study population (q < 0.0001) (Additional file 1). Transcript levels of several inflammatory and lipid metabolism-related genes were regulated after the OGTT (up-regulation with fold changes between 1.11 and 1.36 and down-regulation with fold changes between 0.72 and 0.93 (q < 0.05)) (Additional file 1).
Changes in 2hOGTT gene expression response after dietary intervention
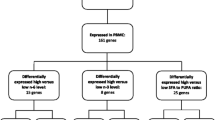
To study if the healthy ND could change the 2hOGTT gene expression response in PBMCs, we conducted a linear multiple regression analysis adjusting for changes at baseline and differences in the study centers. Among the 44 genes, the healthy ND significantly down-regulated the expressions of toll-like receptor 4 (TLR4) (β = −0.33, q = 0.042), interleukin 18 (IL18) (β = −0.73, q = 0.042), and CD36 (β = −0.23, q = 0.042) compared to the CD after the OGTT (Table 3 and Fig. 2a–c). In contrast, a healthy ND significantly up-regulated the expression of peroxisome proliferator-activated receptor delta (PPARD) (β = 0.21, q = 0.042) compared to the CD after the OGTT (Table 3 and Fig. 2d).
Effect of ND compared to CD on gene expression changes after a 2hOGTT. The effect of the healthy ND compared to the CD on TLR4 mRNA expression (a), IL18 mRNA expression (b), CD36 mRNA expression (c), and PPARD mRNA expression (d) presented as fold change (2−ΔCt of 2hOGTT/2−ΔCt of 0hOGTT) in the healthy ND group (n = 42–49) and in the CD group (n = 32–40) after intervention. Fold changes (2−∆ΔCt) are normalized for the reference gene TBP and fasting values (0hOGTT). The box plots show the 2−∆ΔCt values at the end of the intervention. Box plots show the medians with 25th and 75th percentiles. Whiskers express the 1.5 × interquartile range. The number sign is a q value of 0.042. The q values indicate the effect of the ND compared to the CD on gene expression changes after a 2hOGTT. The effect of the independent variable study group is adjusted for fold change at baseline (log2 transformed) and study centers. A q value <0.05 (FDR < 5 %) was considered significant
Discussion
In the present study, we found that the healthy ND modulated the mRNA levels of TLR4, IL18, CD36, and PPARD differently after the OGTT compared to the CD. We also showed that several genes related to inflammation and lipid metabolism were significantly modulated by an OGTT in PBMCs of subjects with MetS at baseline.
Immune response and lipid metabolism are closely linked in metabolic diseases, and alterations in these responses after a food challenge may play an important role in the prevention and early detection of diseases (van Ommen et al. 2009). The healthy ND down-regulated the expression of TLR4 compared to the CD group after the OGTT. TLR4 is involved in the pro-inflammatory response by regulating nuclear factor kappa B (NFκB) activity (Doyle and O’Neill 2006) and is a key regulator of immune response. Previously, it has been reported that an increased TLR4 mRNA expression in monocytes in individuals with MetS compared to healthy controls (Hardy et al. 2013) and down-regulation of the TLR4 mRNA expression by weight loss are associated with improvement of insulin sensitivity in the individuals with MetS (De Mello et al. 2008). This would indicate that the ND compared to the CD may be less inflammatory and may lead to improvement of insulin sensitivity via TLR4 down-regulation in the present study. We were, however, not able to show any improvement in insulin sensitivity or fasting glucose or insulin concentrations in the main SYSDIET trial (Uusitupa et al. 2013), suggesting that the number of subjects in the original study was too low to see an effect. Anyhow, it may be speculated that the use of PBMC gene expression analysis could serve as a more sensitive model system than measurement of circulating markers of glucose metabolism.
Interestingly, we observed that the healthy ND reduced the IL18 mRNA expression after the OGTT compared to CD. IL18 is a pro-inflammatory cytokine shown to be a strong predictor of cardiovascular events in elderly men with MetS, and the effect is stronger with elevated fasting glucose (Troseid et al. 2009). The mRNA expression of IL18 is also increased in obese individuals, and this increase is correlated with insulin resistance (Ahmad et al. 2013). Thus, the down-regulation of IL18 mRNA after the OGTT by healthy ND in the present study may indicate the impact of the ND on disease development among the individuals with MetS.
CD36 is a scavenger receptor involved in lipid uptake and foam cell formation in macrophages (Silverstein 2009). Intracellular lipids taken up by CD36 activate TLR4, which generate pro-inflammatory signals by activating NFκB (Fessler et al. 2009). A blockage of TLR4 and CD36 in human macrophages reduced secretion of IL1β, IL6, and IL8 and the subsequent foam cell formation (Chavez-Sanchez et al. 2014). We found a down-regulation of CD36 mRNA transcript level after the OGTT in the healthy ND group compared to the CD group. We could speculate that the beneficial effects of ND may be executed also via reduction of the foam cell formation and inflammation indicated by down-regulation of the CD36 mRNA expression in the postprandial state. In contrast, the PPARD gene transcript was increased after the OGTT challenge in the ND group compared to the CD group. PPARD is expressed in several tissues in the body, including macrophages. The expression of PPARD genes regulates lipid metabolism and glucose homeostasis, increases fatty acid oxidation, and decreases inflammation as well as platelet activation (Monsalve et al. 2013). We have previously shown that obese subjects at risk had reduced PBMC gene expression of PPARD compared with metabolically healthy obese and control subjects (Telle-Hansen et al. 2013), and PPARD activation improves multiple metabolic disorders (especially blood lipids) in obese subjects (Riserus et al. 2008).
In accordance to other studies, the expression of PDK4 was down-regulated (Zhang et al. 2014), and several pro-inflammatory genes (TNF, TGFB2, CXCR2, CD40LG, IL1RN, CCR2, IL23R, and MMP9) and lipid metabolism related genes (CD36, ABCG1 and ABCA1) were up-regulated, after the OGTT in the whole study population, confirming the use of PBMC gene expression analysis as a model system to detect metabolic responses after an OGTT (Aljada et al. 2006; Aljada et al. 2004; Griffin et al. 2001). The mRNA level of CPT1A was down-regulated during 2hOGTT. Since CPT1 is involved in oxidation of fatty acids, and the oxidation is suppressed in the presence of an adequate glucose supply (Bonnefont et al. 2004), the reduced expression of CPT1 during 2hOGTT may be explained by increased glucose oxidation and decreased fatty acid oxidation.
The strength of this study is the relatively high number of subjects, and to the best of our knowledge, the current dietary intervention study is the first one to use PBMC gene expression as a tool to examine if diet can modify the OGTT response. We used a well-characterized glucose-regulated gene as a positive control to ensure that changes in mRNA level could be measured 2h after OGTT. The limitation of the study is that we cannot differentiate any specific food components responsible for the effect on the change in 2hOGTT response since we did not focus the intervention on single nutrients but on the whole diet. Our primary aim was however to study the effects of the whole diet, since this approach is closer to real-life situations.
Conclusions
We show that the long-term intake of a healthy ND down-regulates genes involved in inflammation and lipid metabolism in individuals at risk for metabolic diseases and thereby may reduce this unfavorable postprandial response. The results need to be confirmed by further human intervention studies, preferably with meal challenges. In addition, experimental models (e.g., ex vivo cell models or animal model) should be employed to extend our biological and clinical understanding of the data presented here.
Methods
Study design and subjects
The study design and participants have been described in detail elsewhere (Uusitupa et al. 2013). In short, this study was a randomized controlled multicenter study performed in six centers within the Nordic countries [Kuopio and Oulu (Finland), Lund and Uppsala (Sweden), Aarhus (Denmark), and Reykjavik (Iceland)]. The participants were randomized after a 4-week run-in period with habitual diet into a healthy ND group or a CD group for 18–24 weeks. The composition of the diets has been described in detail elsewhere (Uusitupa et al. 2013). The main differences between the diets at the nutrient level were the amount of dietary fiber and salt and the quality of dietary fat. Both the ND and the CD were isocaloric based on the evaluation of the habitual diet calculated from a 4-day food record during the run-in period. The Nordic Nutrition Recommendations (NNR) formed the basis of the ND, and the main emphasis was on food items such as whole-grain products, abundant use of berries, fruit and vegetables, rapeseed oil, three fish meals per week, low-fat dairy products, and avoidance of sugar-sweetened products. The subjects in the CD consumed a diet in accordance to the mean nutrient intake in the Nordic countries. Key products were provided to the study participants in both groups. The study participants were advised to keep body weight and physical activity constant and not to change their smoking and drinking habits or drug treatment during the study. All study participants provided their written informed consent, and local ethics committees of all the participating centers approved the study protocol.
Altogether, 309 individuals were originally contacted and screened at the study clinics, and 213 were randomized as described earlier (Uusitupa et al. 2013). Ninety-six individuals in the ND group and 70 in the CD group completed the trial (Uusitupa et al. 2013). The inclusion criteria were age 30–65 years, BMI 27–38 kg m−2, and two other of the International Diabetes Federation (IDF) criteria for MetS (Alberti et al. 2009). Antihypertensive and lipid-lowering medication, as well as inhaled corticosteroids, were allowed but without dosage changes during the trial. The main exclusion criteria included any chronic disease and condition, which could hamper the adherence to the dietary intervention protocol, poor compliance, chronic liver, thyroid and kidney diseases, alcohol abuse (>40 g per day), diabetes, fasting triglycerides >3.0 mM, total cholesterol >6.5 mM, and blood pressure >160/100 mmHg. A few study participants with triglycerides between 3 and 4 mM and with BMI between 38 and <40 kg m−2 were, however, included in the main study population.
In this present sub-study of the SYSDIET trial, we included a total of 94 subjects (n = 54 in ND and n = 40 in CD) out of the 166 subjects who completed the SYSDIET study (Fig. 3). We excluded subjects from Aarhus (n = 31), Uppsala (n = 9), and Reykjavik (n = 15), because the Aarhus study center did not collect PBMC samples, and the number of PBMC samples was limited from Uppsala (n = 9) and Reykjavik (n = 5). So, by excluding these two centers, we reduced variance. We also excluded subjects with high-sensitivity C-reactive protein (hs-CRP) concentration higher than 10 mg L−1 at baseline or after the intervention (n = 6), baseline BMI above 39 kg m−2 (n = 3), and body weight change more than 4 kg during the intervention (n = 8). Since five subjects were excluded from the analysis due to low quantity of RNA (n = 3) or problems with the TaqMan Array Micro Fluidic Cards (n = 2), we analyzed data from 89 subjects (n = 49 in ND and n = 40 in CD) (Fig. 3).
Clinical and biochemical measurements
Procedures regarding the clinical and biochemical measurements have been described previously (Uusitupa et al. 2013). In short, subjects were examined in the morning after overnight fasting. Anthropometric measurements were performed locally according to the standard operational procedures. Concentrations of plasma glucose, cholesterol, and triglycerides were analyzed locally in the centers using routine methodology. Blood samples to measure cytokines and adipokines from all the study centers were analyzed in the University of Eastern Finland and Kuopio University Hospital, Finland. Plasma insulin was analyzed in the Aarhus University Hospital, Denmark, using routine automated clinical chemistry analyzers.
Standard 2hOGTT
A standard 2hOGTT (75 g D-glucose) was performed after an overnight fast at baseline and at the end of the intervention. Blood samples for PBMC isolation were taken at the time points 0 and 120 min.
PBMCs, RNA isolation, cDNA synthesis, and qPCR
After blood collection, PBMCs were isolated at baseline and at the end of the intervention at time points 0 and 120 min by using the BD Vacutainer Cell Preparation tubes according to the manufacturer’s instructions (Becton, Dickinson San Jose, CA, USA) and stored as pellets at −80 °C for further analysis. Total RNA isolation was performed centrally at the Karolinska Institute (Stockholm, Sweden). The total RNA was isolated using the RNeasy Mini Kit according to the manufacturer’s instructions (Qiagen, Valencia, CA, USA). RNA quantity and quality measurements were performed using a Nanodrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Gothenburg, Sweden) and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), respectively. RNA from all samples was reverse transcribed by a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA). Quantitative real-time polymerase chain reaction (qPCR) was performed on an ABI PRISM 7900HT (Applied Biosystems) using TaqMan Array Micro Fluidic Cards (Applied Biosystems). The target genes are shown in the Additional file 1. Primer sequences are commercially available (Applied Biosystems) and can be provided upon request. The selection of target genes was primarily based on previous long-term and short-term dietary intervention studies where PBMC gene expression of inflammatory and lipid metabolism genes was modulated or associated with features of MetS or metabolic risk factors for T2DM and CVD (De Mello et al. 2009; Bouwens et al. 2009; Jones et al. 2011; Kaminski et al. 1993) and on results from our own studies (Myhrstad et al. 2011; Telle-Hansen et al. 2013).
The relative mRNA level for each transcript was calculated by the ∆Δ cycle threshold (Ct) method (Livak and Schmittgen 2001). TATA-box binding protein (TBP) was used as reference gene for normalization. Briefly, the Ct values of each target gene were normalized to the Ct values of the TBP (=ΔCt). The fold change in mRNA gene expression from TBP was calculated at fasting (0h) and after (2h) OGTT (2−ΔCt) at baseline and at the end of the intervention. The fold change in mRNA gene expression from fasting was calculated as 2−∆ΔCt at baseline and at the end of the intervention, as 2−ΔCt 2hOGTT was divided by 2−ΔCt 0hOGTT.
Statistical analysis
For baseline characteristic comparisons, we used independent t test to test the difference between means, Mann-Whitney U test to test the difference between medians and chi-square test to test the difference between categorical variables. Power calculations (alpha <0.05, beta >0.8) were carried out on serum cholesterol, fasting glucose, and insulin (Uusitupa et al. 2013).
Linear multivariable regression analyses were used to test the independent effect of the study groups on the change in glucose, insulin, triglycerides, and free fatty acids from 0h (fasting) to 2h (after OGTT) at the end of the intervention as well as on the dependent dietary intake variables at the end of the intervention. The effect of the independent variable study group was adjusted for the corresponding baseline variable, study centers, gender, log10-transformed age, and body weight at the end of the intervention. Changes in gene expression from 0h to 2hOGTT within the whole study population were tested with Wilcoxon signed rank test (2−ΔCt). Data are given as the median (25–75th percentiles). Linear multivariable regression analyses were also carried out to test the independent effect of the study groups on the dependent variable fold change at the end of the intervention. The analyses were adjusted for the independent variables fold change at baseline and study center. In the presentation, β denotes the regression coefficient of the treatment group. Fold changes at baseline and at the end of the intervention (2−∆ΔCt) were log2 transformed before the analyses to improve normality. To account for multiple testing, we applied false discovery rate (FDR) analysis and q < 0.05 (FDR < 5 %) was considered significant. Calculations were performed using IBM SPSS Statistics version 20 (Armonk, NY, USA) and R version 3.2.0.
Abbreviations
- ABCA1:
-
ATP-binding cassette, sub-family A (ABC1), member 1
- ABCG1:
-
ATP-binding cassette, sub-family G, member 1
- CCL2:
-
chemokine (C-C motif) ligand 2
- CCL5:
-
chemokine (C-C motif) ligand 5
- CCR2:
-
chemokine (C-C motif) receptor 2
- CCR4:
-
chemokine (C-C motif) receptor 4
- CD:
-
control diet
- CD36:
-
CD36 molecule (thrombospondin receptor)
- CD40:
-
CD40 molecule
- CD40LG:
-
CD40 ligand
- CPT1A:
-
carnitine palmitoyltransferase 1A
- CPT1B:
-
carnitine palmitoyltransferase 1B
- CRAT:
-
carnitine O-acetyltransferase
- CXCR2:
-
chemokine (C-X-C motif) receptor 2
- CVD:
-
cardiovascular diseases
- FDR:
-
false discovery rate
- HMGCR:
-
3-hydroxy-3-methylglutaryl-CoA reductase
- ICAM1:
-
intercellular adhesion molecule 1
- IFNG:
-
interferon, gamma
- IKBKB:
-
inhibitor of kappa light polypeptide gene enhancer in B cells, kinase beta
- IL18:
-
interleukin 18
- IL1B:
-
interleukin 1, beta
- IL1RN:
-
interleukin 1 receptor antagonist
- IL23A:
-
interleukin 23, alpha subunit p19
- IL23R:
-
interleukin 23, receptor
- IL6:
-
interleukin 6
- IL8:
-
interleukin 8
- LDLR:
-
low-density lipoprotein receptor
- LIPE:
-
lipase, hormone-sensitive
- MetS:
-
metabolic syndrome
- MMP9:
-
matrix metallopeptidase 9
- NAMPT:
-
nicotinamide phosphoribosyltransferase
- ND:
-
Nordic diet
- NFKBIA:
-
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha
- OLR1:
-
oxidized low density lipoprotein (lectin-like) receptor 1
- PBMCs:
-
peripheral blood mononuclear cells
- PDGFA:
-
platelet-derived growth factor alpha polypeptide
- PDGFB:
-
platelet-derived growth factor beta polypeptide
- PDK4:
-
pyruvate dehydrogenase kinase, isozyme 4
- PLIN2:
-
perilipin 2
- PPARA:
-
peroxisome proliferator-activated receptor alpha
- PPARD:
-
peroxisome proliferator-activated receptor delta
- RELA:
-
v-rel reticuloendotheliosis viral oncogene homologue A
- SREBF1:
-
sterol regulatory element binding transcription factor 1
- SYSDIET:
-
Systems Biology in Controlled Dietary Interventions and Cohort Studies
- T2DM:
-
type 2 diabetes mellitus
- TGFB2:
-
transforming growth factor beta 2
- TLR4:
-
toll-like receptor 4
- TNF:
-
tumor necrosis factor
- TNFRSF1A:
-
tumor necrosis factor receptor superfamily member 1A
- TNFRSF1B:
-
tumor necrosis factor receptor superfamily member 1B
- UCP2:
-
uncoupling protein 2
References
Adamsson V, Reumark A, Fredriksson IB, Hammarstrom E, Vessby B, Johansson G, Riserus U (2011) Effects of a healthy Nordic diet on cardiovascular risk factors in hypercholesterolaemic subjects: a randomized controlled trial (NORDIET). J Intern Med 269:150–159
Afman L, Milenkovic D, Roche HM (2014) Nutritional aspects of metabolic inflammation in relation to health-insights from transcriptomic biomarkers in PBMC of fatty acids and polyphenols. Molecular nutrition & food research
Ahmad R, Al-Mass A, Al-Ghawas D, Shareif N, Zghoul N, Melhem M, Hasan A, Al-Ghimlas F, Dermime S, Behbehani K (2013) Interaction of osteopontin with IL-18 in obese individuals: implications for insulin resistance. PLoS One 8:e63944
Alberti KG, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, Fruchart JC, James WP, Loria CM, Smith SC Jr (2009) Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation 120:1640–1645
Aljada A, Ghanim H, Mohanty P, Syed T, Bandyopadhyay A, Dandona P (2004) Glucose intake induces an increase in activator protein 1 and early growth response 1 binding activities, in the expression of tissue factor and matrix metalloproteinase in mononuclear cells, and in plasma tissue factor and matrix metalloproteinase concentrations. Am J Clin Nutr 80:51–57
Aljada A, Friedman J, Ghanim H, Mohanty P, Hofmeyer D, Chaudhuri A, Dandona P (2006) Glucose ingestion induces an increase in intranuclear nuclear factor kappaB, a fall in cellular inhibitor kappaB, and an increase in tumor necrosis factor alpha messenger RNA by mononuclear cells in healthy human subjects. Metabolism 55:1177–1185
Bastard JP, Maachi M, Lagathu C, Kim MJ, Caron M, Vidal H, Capeau J, Feve B (2006) Recent advances in the relationship between obesity, inflammation, and insulin resistance. Eur Cytokine Netw 17:4–12
Bonnefont JP, Djouadi F, Prip-Buus C, Gobin S, Munnich A, Bastin J (2004) Carnitine palmitoyltransferases 1 and 2: biochemical, molecular and medical aspects. Mol Asp Med 25:495–520
Bouwens M, van de Rest O, Dellschaft N, Bromhaar MG, De Groot LC, Geleijnse JM, Muller M, Afman LA (2009) Fish-oil supplementation induces antiinflammatory gene expression profiles in human blood mononuclear cells. Am J Clin Nutr 90:415–424
Bouwens M, Grootte BM, Jansen J, Muller M, Afman LA (2010) Postprandial dietary lipid-specific effects on human peripheral blood mononuclear cell gene expression profiles. Am J Clin Nutr 91:208–217
Brader L, Uusitupa M, Dragsted LO, Hermansen K (2014) Effects of an isocaloric healthy Nordic diet on ambulatory blood pressure in metabolic syndrome: a randomized SYSDIET sub-study. Eur J Clin Nutr 68:57–63
Chavez-Sanchez L, Garza-Reyes MG, Espinosa-Luna JE, Chavez-Rueda K, Legorreta-Haquet MV, Blanco-Favela F (2014) The role of TLR2, TLR4 and CD36 in macrophage activation and foam cell formation in response to oxLDL in humans. Hum Immunol 75:322–329
Choi HJ, Yun HS, Kang HJ, Ban HJ, Kim Y, Nam HY, Hong EJ, Jung SY, Jung SE, Jeon JP, Han BG (2012) Human transcriptome analysis of acute responses to glucose ingestion reveals the role of leukocytes in hyperglycemia-induced inflammation. Physiol Genomics 44:1179–1187
Cruz-Teno C, Perez-Martinez P, Delgado-Lista J, Yubero-Serrano EM, Garcia-Rios A, Marin C, Gomez P, Jimenez-Gomez Y, Camargo A, Rodriguez-Cantalejo F et al (2012) Dietary fat modifies the postprandial inflammatory state in subjects with metabolic syndrome: the LIPGENE study. Mol Nutr Food Res 56:854–865
De Mello VD, Kolehmainen M, Pulkkinen L, Schwab U, Mager U, Laaksonen DE, Niskanen L, Gylling H, Atalay M, Rauramaa R, Uusitupa M (2008) Downregulation of genes involved in NFkappaB activation in peripheral blood mononuclear cells after weight loss is associated with the improvement of insulin sensitivity in individuals with the metabolic syndrome: the GENOBIN study. Diabetologia 51:2060–2067
De Mello VD, Erkkila AT, Schwab US, Pulkkinen L, Kolehmainen M, Atalay M, Mussalo H, Lankinen M, Oresic M, Lehto S, Uusitupa M (2009) The effect of fatty or lean fish intake on inflammatory gene expression in peripheral blood mononuclear cells of patients with coronary heart disease. Eur J Nutr 48:447–455
Doyle SL, O’Neill LA (2006) Toll-like receptors: from the discovery of NFkappaB to new insights into transcriptional regulations in innate immunity. Biochem Pharmacol 72:1102–1113
Fessler MB, Rudel LL, Brown JM (2009) Toll-like receptor signaling links dietary fatty acids to the metabolic syndrome. Curr Opin Lipidol 20:379–385
Griffin E, Re A, Hamel N, Fu C, Bush H, McCaffrey T, Asch AS (2001) A link between diabetes and atherosclerosis: Glucose regulates expression of CD36 at the level of translation. Nat Med 7:840–846
Hardy OT, Kim A, Ciccarelli C, Hayman LL, Wiecha J (2013) Increased Toll-like receptor (TLR) mRNA expression in monocytes is a feature of metabolic syndrome in adolescents. Pediatric Obesity 8:e19–e23
Jones JL, Park Y, Lee J, Lerman RH, Fernandez ML (2011) A Mediterranean-style, low-glycemic-load diet reduces the expression of 3-hydroxy-3-methylglutaryl-coenzyme A reductase in mononuclear cells and plasma insulin in women with metabolic syndrome. Nutr Res 31:659–664
Kaminski WE, Jendraschak E, Kiefl R, Von SC (1993) Dietary omega-3 fatty acids lower levels of platelet-derived growth factor mRNA in human mononuclear cells. Blood 81:1871–1879
Kempf K, Rose B, Herder C, Haastert B, Fusbahn-Laufenburg A, Reifferscheid A, Scherbaum WA, Kolb H, Martin S (2007) The metabolic syndrome sensitizes leukocytes for glucose-induced immune gene expression. J Mol Med (Berl) 85:389–396
Laaksonen DE, Toppinen LK, Juntunen KS, Autio K, Liukkonen KH, Poutanen KS, Niskanen L, Mykkanen HM (2005) Dietary carbohydrate modification enhances insulin secretion in persons with the metabolic syndrome. Am J Clin Nutr 82:1218–1227
Libby P, Ridker PM, Hansson GK (2009) Inflammation in atherosclerosis: from pathophysiology to practice. J Am Coll Cardiol 54:2129–2138
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods 25:402–408
Monsalve FA, Pyarasani RD, Delgado-Lopez F, Moore-Carrasco R (2013) Peroxisome proliferator-activated receptor targets for the treatment of metabolic diseases. Mediat Inflamm 2013:549627
Mozaffarian D, Appel LJ, Van HL (2011) Components of a cardioprotective diet: new insights. Circulation 123:2870–2891
Myhrstad MC, Narverud I, Telle-Hansen VH, Karhu T, Lund DB, Herzig KH, Makinen M, Halvorsen B, Retterstol K, Kirkhus B et al (2011) Effect of the fat composition of a single high-fat meal on inflammatory markers in healthy young women. Br J Nutr 106:1826–1835
Myhrstad MC, Ulven SM, Gunther CC, Ottestad I, Holden M, Ryeng E, Borge GI, Kohler A, Bronner KW, Thoresen M, Holven KB (2014) Fish oil supplementation induces expression of genes related to cell cycle, endoplasmic reticulum stress and apoptosis in peripheral blood mononuclear cells: a transcriptomic approach. J Intern Med
O’Grada CM, Morine MJ, Morris C, Ryan M, Dillon ET, Walsh M, Gibney ER, Brennan L, Gibney MJ, Roche HM (2014) PBMCs reflect the immune component of the WAT transcriptome—implications as biomarkers of metabolic health in the postprandial state. Mol Nutr Food Res 58:808–820
Paniagua JA, de la Sacristana AG, Sanchez E, Romero I, Vidal-Puig A, Berral FJ, Escribano A, Moyano MJ, Perez-Martinez P, Lopez-Miranda J, Perez-Jimenez F (2007) A MUFA-rich diet improves postprandial glucose, lipid and GLP-1 responses in insulin-resistant subjects. J Am Coll Nutr 26:434–444
Riserus U, Sprecher D, Johnson T, Olson E, Hirschberg S, Liu A, Fang Z, Hegde P, Richards D, Sarov-Blat L et al (2008) Activation of peroxisome proliferator-activated receptor (PPAR)delta promotes reversal of multiple metabolic abnormalities, reduces oxidative stress, and increases fatty acid oxidation in moderately obese men. Diabetes 57:332–339
Silverstein RL (2009) Inflammation, atherosclerosis, and arterial thrombosis: role of the scavenger receptor CD36. Cleve Clin J Med 76(Suppl 2):S27–S30
Telle-Hansen VH, Halvorsen B, Dalen KT, Narverud I, Wesseltoft-Rao N, Granlund L, Ulven SM, Holven KB (2013) Altered expression of genes involved in lipid metabolism in obese subjects with unfavourable phenotype. Genes Nutr 8:425–434
Troseid M, Seljeflot I, Hjerkinn EM, Arnesen H (2009) Interleukin-18 is a strong predictor of cardiovascular events in elderly men with the metabolic syndrome: synergistic effect of inflammation and hyperglycemia. Diabetes Care 32:486–492
Uusitupa M, Hermansen K, Savolainen MJ, Schwab U, Kolehmainen M, Brader L, Mortensen LS, Cloetens L, Johansson-Persson A, Onning G et al (2013) Effects of an isocaloric healthy Nordic diet on insulin sensitivity, lipid profile and inflammation markers in metabolic syndrome—a randomized study (SYSDIET). J Intern Med 274:52–66
van Dijk SJ, Feskens EJ, Bos MB, De Groot LC, De Vries JH, Muller M, Afman LA (2012a) Consumption of a high monounsaturated fat diet reduces oxidative phosphorylation gene expression in peripheral blood mononuclear cells of abdominally overweight men and women. J Nutr 142:1219–1225
van Dijk SJ, Mensink M, Esser D, Feskens EJ, Muller M, Afman LA (2012b) Responses to high-fat challenges varying in fat type in subjects with different metabolic risk phenotypes: a randomized trial. PLoS One 7:e41388
van Ommen B, Keijer J, Heil SG, Kaput J (2009) Challenging homeostasis to define biomarkers for nutrition related health. Mol Nutr Food Res 53:795–804
Visvikis-Siest S, Marteau JB, Samara A, Berrahmoune H, Marie B, Pfister M (2007) Peripheral blood mononuclear cells (PBMCs): a possible model for studying cardiovascular biology systems. Clin Chem Lab Med 45:1154–1168
Wellen KE, Hotamisligil GS (2005) Inflammation, stress, and diabetes. J Clin Invest 115:1111–1119
Zhang S, Hulver MW, McMillan RP, Cline MA, Gilbert ER (2014) The pivotal role of pyruvate dehydrogenase kinases in metabolic flexibility. Nutr Metab 11:10
Acknowledgements
We thank Maritta Siloaho (MS) for the excellent expertise and advice for biochemical measurements and Marika Rönnholm (MR), Kaija Kettunen (KK), Tuomas Onnukka (TO), and Erja Kinnunen (EK) for the excellent technical assistance.
Funding
The project was funded by NordForsk Nordic Centre of Excellence in Food, Nutrition and Health project 070014 [SYSDIET (Systems Biology in Controlled Dietary Interventions and Cohort Studies)] and further, by Oslo and Akershus University College of Applied Sciences (Norway), Academy of Finland (131593 to VDdM), University of Oslo (Norway), Swedish Research council, Svenska Diabetesförbundet, SRP Diabetes, Finnish Diabetes Research Foundation, Finnish Foundation for Cardiovascular Research, the Sigrid Juselius Foundation, EVO funding from Kuopio University Hospital (Finland), the Druvan Foundation, Skåne University Hospital, the Heart-Lung Foundation, Diabetesfonden and Foundation Cerealia (Sweden), the Danish Obesity Research Centre (DanORC, www.danorc.dk), the Danish Council for Strategic Research (DairyHealth, BioFunCarb) (Denmark), the Agricultural Productivity Fund, and the Research Fund of the University of Iceland (Iceland).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MK, ID, MCWM, VdM, JP, CC, US, K-HH, LC, MUS, JH, MJS, FR, KH, LOD, IG, IT, UR, BA, PA, KSP, MU, and SMU designed the research. LL, MK, IN, ID, US, K-HH, LC, MUS, JH, MJS, FR, KH, LOD, IG, IT, UR, BA, PA, MU, KBH, and SMU conducted the research. LL, MK, IN, MT, KBH, and SMU analyzed the data or performed the statistical analysis. LL, MK, KBH, and SMU wrote the first draft of the manuscript and had primary responsibility for the final content. MK, KSP, and MU were responsible for the coordination of the SYSDIET consortium. All authors have participated in and critically reviewed the manuscript and accepted it to be submitted.
Additional file
Additional file 1:
mRNA level at fasting (0hOGTT) and after 2hOGTT and fold change from fasting in the whole study population at baseline. Data for 0hOGTT and 2hOGTT is given as 2−∆Ct (normalized for TBP). Data for fold change is given as 2−∆∆Ct (normalized for TBP and 0hOGTT values). All values are presented as medians with 25th–75th percentiles. (DOCX 21.4 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Leder, L., Kolehmainen, M., Narverud, I. et al. Effects of a healthy Nordic diet on gene expression changes in peripheral blood mononuclear cells in response to an oral glucose tolerance test in subjects with metabolic syndrome: a SYSDIET sub-study. Genes Nutr 11, 3 (2016). https://doi.org/10.1186/s12263-016-0521-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12263-016-0521-4