Abstract
The incidence and mortality of colorectal cancer (CRC) are rapidly increasing worldwide. Recently, there has been significant attention given to N6-methyladenosine (m6A), the most common mRNA modification, especially for its effects on CRC development. It is important to note that the progression of CRC would be greatly hindered without the tumor microenvironment (TME). The interaction between CRC cells and their surroundings can activate and influence complex signaling mechanisms of epigenetic changes to affect the survival of tumor cells with a malignant phenotype. Additionally, the TME is influenced by m6A regulatory factors, impacting the progression and prognosis of CRC. In this review, we describe the interactions and specific mechanisms between m6A modification and the metabolic, hypoxia, inflammatory, and immune microenvironments of CRC. Furthermore, we summarize the therapeutic role that m6A modification can play in the CRC microenvironment, and discuss the current status, limitations, and potential future directions in this field. This review aims to provide new insights into the molecular targets and theoretical foundations for the treatment of CRC.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is a common malignancy of the digestive system, with its incidence and mortality increasing worldwide (Arnold et al. 2017). It ranks as the third deadliest tumor and the fourth most frequently diagnosed tumor in the world (Sung et al. 2021). Despite advancements in technology and treatment options, the estimated mortality of CRC patients remains high (Cao et al. 2021a). Various risk factors contribute to the development of CRC, including diet, intestinal metabolism, genetics, polyp lesions, and chronic inflammation. These factors are interconnected and cannot exist independently. For instance, a high-fat diet (HFD) not only stimulates the proliferation of intestinal mucosal and cancer cells but also inhibits the proliferation of lymphocytes in the lamina propria and weakens immune function. This leads to intestinal barrier dysfunction and dysregulation of intestinal metabolism (Yang et al. 2022; Ocvirk and O'Keefe 2021). Disturbances in intestinal metabolism further promote chronic inflammation and CRC through various enzymes and metabolites (Jackson and Theiss 2020).
The tumor microenvironment (TME) is a dynamic environment that plays a crucial role in tumor cell survival, growth, proliferation, and metastasis (Feingold et al. 1997). It consists of a complex and heterogeneous system comprising both cellular and non-cellular elements. The cellular components of the TME include cancer-associated fibroblasts (CAFs), T lymphocytes, B lymphocytes, NK cells, tumor-associated macrophages (TAMs), tumor-associated neutrophils (TANs), and endothelial cells (Arneth 2019). In addition, there are non-cellular components such as growth factors, cytokines, and extracellular matrix that surround the tumor cells. These components interact and form specific metabolic, hypoxia, inflammatory, and immune microenvironments in CRC. They are responsible for the regulation of various pro- and anti-tumor factors and play a role in the progression of the disease. The composition of TME is influenced by various signaling pathways and cytokine flow, which involve a variety of chemical modifications. These modifications include ubiquitination, 5-methylcytosine (m5C), N6-methyladenosine (m6A), and 7-methylguanosine (m7G) modifications. Among them, m6A modification has been the most extensively studied in recent years (Jordan et al. 2018), (Maybin et al. 2018), (Catana et al. 2015), (Wu and Dai 2017).
Recently, m6A modification has emerged as a new regulatory mechanism in eukaryotes and is one of the most common RNA methylation modifications. It is a reversible epigenetic modification that occurs in both mRNAs and non-coding RNAs (ncRNAs). The m6A modification is catalyzed by certain methyltransferases. Following the m6A modification, methylated binding proteins precisely identify and bind to the modified RNA. These modifications can be reversed, and demethylases are responsible for their removal (Han et al. 2023). By modulating these processes, m6A modifications have a profound impact on the fate and cellular functions of the modified RNA molecules, influencing RNA splicing, export, translation, and stability. Furthermore, they play a crucial role in almost all essential biological processes, including the malignant progression of tumors (Deng et al. 2018; Alarcon et al. 2015). The progression of tumors is highly dependent on the microenvironment in which they are located. Several recent studies have attempted to uncover the correlation between m6A and the TME (Han et al. 2019a; Wang et al. 2020a; Li et al. 2021a). The m6A methylation recognition protein YT521-B homology domain family protein 1 (YTHDF1) regulates the tumor immune microenvironment. Deletion of YTHDF1 enhances the anti-tumor activity of CD8+T cells and inhibits the translation efficiency of lysosomal histone proteases in dendritic cells (DCs) (Han et al. 2019a). Overexpression of the m6A methylesterase Methyltransferase-like 3 (METTL3) alters the metabolic microenvironment of gastric cancer and promotes malignant tumor progression and liver metastasis (Wang et al. 2020a). Furthermore, components of the tumor microenvironment have been found to regulate the expression of m6A methylation regulators. For example, the hypoxia-inducible factor-1α (HIF-1α) can regulate the abundance of m6A (Li et al. 2021a). These findings suggest that the tumor microenvironment also plays a role in the complex regulatory network of m6A modification. The interactions between m6A and the tumor microenvironment are critical for tumor progression.
Although the correlation between m6A modification and the TME has been extensively studied, there is a scarcity of insights into the variants, functional characteristics, TME associations, and related clinical implications of m6A regulators in CRC. This review aims to summarize and demonstrate the specific roles of m6A regulators in the metabolic, hypoxic, inflammatory, and immune microenvironments associated with CRC, based on their reciprocal regulation with the TME. Additionally, our review highlights the potential diagnostic and therapeutic value of m6A modifications in the TME, discusses current research gaps, and suggests novel directions for future investigations.
Molecular composition of m6A
The molecular composition of m6A includes methyltransferase, demethylase, and recognition factor, also termed m6A “writers”, “erasers” and “readers” respectively. These proteins have the ability to add, delete, or recognize m6A modification sites, and impacting crucial biological processes. Any factor that influences the expression levels of “writers” and “erasers” will consequently affect the activity of m6A in cells, resulting in abnormal levels of m6A in tumors. On the other hand, “readers” play dominant role in post-modification regulation of target mRNAs (An and Duan 2022). The mechanisms by which m6A enzymes recognize and regulate mRNA levels of specific target proteins are as follows: (a) certain “readers” recruit eukaryotic initiation translation factors and bind m6A labeled mRNA to ribosomes,(b) specific transcripts are bound by certain “readers,” thereby affecting mRNA translation; (c) some “writers” directly interact with transcription factors to mediate mRNA cyclization; (d) through histone modification, some “writers” are recruited to specific mRNA regions (Liu et al. 2023). These m6A enzymes regulate the expression level of proto-oncogenes or tumor suppressor genes by influencing the transcription, maturation, translation and degradation of RNA, ultimately participating in the occurrence and development of tumors (He et al. 2019).
METTL3 was the first characteristic component of m6A "writers" to be identified (Bokar et al. 1997). Currently, it is believed to primarily function as an oncogene, promoting tumor progression by adding m6A modifications to key transcripts (Li et al. 2019). METTL14 is another active component of m6A "writers" that binds to METTL3, forming stable heterodimeric complexes (Liu et al. 2014). While METTL3 is the catalytically active subunit, METTL14 plays a crucial role in recognizing the substrate structure (Wang et al. 2016). The third active component of m6A “writers” is Wilms’ tumor 1-associated protein (WTAP), which lacks catalytic activity but is essential for the nuclear localization and interaction between METTL3 and METTL14 (Scholler et al. 2018). METTL16, a homolog of METTL3, regulates U6 small nuclear RNA (U6 snRNA) methylation (Ruszkowska 2021). As a protective gene, the expression level of METTL16 has been found to exhibit a positively correlated with overall survival of several cancers (Li et al. 2020a). Conversely, a distinct investigation revealed that elevated expression of METTL16 was associated with a poor survival rate in patient with breast cancer (Zhang et al. 2020). Additionally, other components of m6A “writers” include virlike m6A methyltransferase associated (VIRMA/KIAA1429), RNA-binding motif protein 15 (RBM15), Cbl proto-oncogene, E3 ubiquitin-protein ligase-like 1 (CBLL1) and zinc finger CCCH-Type containing 13 (ZC3H13) (Jiang et al. 2021). VIRMA/KIAA1429 has been implicated in the malignant proliferation of CRC cells (Li et al. 2023a). CBLL1 is involved in the development of inflammatory bowel disease, and its dysregulation is associated with the inflammatory microenvironment of CRC (Roca-Lema et al. 2022). ZC3H13 has been demonstrated to suppress CRC invasion and proliferation by deactivating the Ras-ERK signaling pathway (Zhu et al. 2019).
In the current study, the m6A “erasers” consist of three main proteins: fat mass and obesity-associated protein (FTO), α-ketoglutarate-dependent dioxygenase homolog 5 (ALKBH5), and ALKBH3 (Huang et al. 2021; Dai et al. 2018). Jia et al. were the first to identify the demethylation activity of FTO in vitro against abundant m6A residues in RNA, indicating the reversible nature of m6A modification and its dynamic regulation (Jia et al. 2011).
Due to the multiple tumors signaling pathways FTO interacts with, the expression and effects of FTO in various tumors, whether it promotes or suppresses tumor growth, remain a subject of controversy. ALKBH5 was proven to have demethylation activity for the first time in 2013 (Zheng et al. 2013). Both ALKBH5 and FTO belong to the α-ketoglutarate-dependent dioxygenase family and catalyze m6A demethylation in an α-ketoglutarate-dependent manner (Marcinkowski et al. 2020). ALKBH3, a recently discovered m6A demethylase, regulates tumor progression by inducing tRNA demethylation and the production of corresponding miRNAs and proteins (Chen et al. 2019).
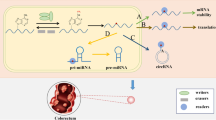
The m6A “readers” activate downstream effector proteins or complexes in the trans conformation through recognition, which is an essential part of the biological function of m6A modifications (Yang et al. 2018; Allis and Jenuwein 2016). YT521-B homology domain family proteins 1–3 (YTHDF1-3) are three parallel sequences of the YTHDF family and they all have structural domains that selectively bind to m6A and are responsible for enhancing translation and degradation of mRNAs (Xiao et al. 2015; Li et al. 2017a). YTH domain-containing protein 1 (YTHDC1) promotes exon inclusion by recruiting the pri-mRNA splicing factor SRSF3 and blocking SRSF10 from binding to mRNA (Xiao et al. 2016). YTH domain-containing protein 2 (YTHDC2) can preferentially bind m6A-modified mRNA and affect its stability (Lma et al. 2020). In addition, several members of the heterogeneous nuclear ribonucleoprotein (HNRNP) family have also been shown to act as m6A "readers", such as HNRNPA2B1 and HNRNPC. They can regulate miRNA or mRNA abundance by processing m6A-modified RNA transcripts (Alarcón et al. 2015b; Lv et al. 2021). Moreover, insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) (including IGF2BP1/2/3), and the IGF-II mRNA-binding proteins (IMPs) family are also considered as m6A “readers” that can recognize specific m6A sequences targeted to mRNA for transcription (Huang et al. 2018). Figure 1 illustrates the classification and composition of m6A regulators.
Classification and composition of m6A regulators. m6A “writers” include METTL3, METTL14, WTAP, METTL16, KIAA1429, RBM15, RBM15B, CBLL1, ZC3H13, which are responsible for structure stabilization and catalysis. m6A “erasers” include FTO, ALKBH5, ALKBH3, which are responsible for demethylation. HNRNPA2B1, HNRNPC, and IGF2BPs are responsible for recognizing m6A sites and mediating RNA splicing, translation, processing, structural transformation, and stabilization
m6A modification and TME of CRC
Metabolic microenvironment
The progression of CRC is closely related to the intestinal metabolic microenvironment, which includes factors such as intestinal flora, glucose metabolism, and lipid metabolism. It is believed that consuming high levels of dietary fiber and maintaining a stable intestinal metabolism can reduce the risk of CRC (Slavin 2008).
Recent study has revealed that the m6A methyltransferase METTL3 is responsible for regulating the cell cycle protein E1 (CCNE1) in CRC cells (Zhu et al. 2020). METTL3 promotes CRC proliferation by methylating the m6A site on CCNE1 mRNA. However, the intestinal microbial metabolite butyrate reduces m6A levels in CRC cells, reversing this process. Conversely, overexpression of METTL3 can counteract the inhibitory effects of butyrate on CRC progression (Zhu et al. 2020). In addition, the presence of Enterotoxigenic Bacteroides fragilis (ETBF) can downregulate miR-149-3p, leading to intestinal inflammation and promoting CRC. This process is dependent on METTL14-mediated m6A modification (Cao et al. 2021b). These finding suggest that there is a reciprocal relationship between the gut microbial metabolic environment and m6A regulators, which is closely linked to CRC progression. Another important factor in CRC is the protein KIAA1429, which targets HK2 mRNA to accelerate aerobic glycolysis and the production of malignant phenotypes in CRC (Li et al. 2022a). METTL3 also induces glycolysis and enhances CRC proliferation by promoting PTTG3P, which relies on IGF2BP2 for recognition its m6A binding site (Zheng et al. 2021). IMP2, a member of the m6A "readers", modifies the RNA metabolism of ZFAS1 through m6A modification. This process influences the energy metabolism of cell mitochondria, promotes CRC proliferation, and suppresses apoptosis of CRC cells (Lu et al. 2021). Concerning lipid metabolism, significant alterations have been observed in CRC patients. The enzyme DEGS2, which plays a crucial role in lipid metabolism, promotes CRC proliferation and metastasis in an m6A-dependent manner (Guo et al. 2021). In addition, chemoresistance is a major factor contributing to the failure of CRC chemotherapy. Drug-resistant cells undergo metabolic reprogramming to regulate the metabolic microenvironment within CRC. METTL3 has been shown to regulate glucose metabolism in CRC by enhancing the expression of LDHA and mediating resistance of CRC cells to 5-FU (Zhang et al. 2022a).
Together, m6A modifications play a bridging role that links the metabolic microenvironment to the progression of CRC. Certain components of the metabolic microenvironment in which CRC resides can influence its progression by affecting m6A modification. Current study specifically focuses on the interaction between intestinal microbe and m6A modification. Some of the intestinal flora implicated in CRC, such ads Enterococcus faecalis and Helicobacter hepaticus, are also responsible for producing numerous metabolites. However, whether these components are directly related to m6A modification requires further investigation. In the case of the glycolipid metabolic environment of CRC, m6A modifications only regulate this process in one direction. It is still unknown whether the metabolic pattern and the metabolites themselves also affect m6A modification in CRC. Figure 2 illustrates the role of m6A regulators in the metabolic microenvironment of CRC.
m6A modifications interact with the CRC metabolic microenvironment. m6A “writers” METTL3 promotes CRC proliferation by targeting LDHA and PTTG3P to regulate glycolysis. KIAA1429 promotes CRC proliferation by increasing the expression of HK2. METTL14 promotes CRC progression by inhibiting miR-149-3p. m6A “readers”, IPM2 targets ZFAS1 to mediate mitochondrial metabolism and inhibit CRC proliferation. YTHDF1 promotes CRC chemoresistance by mediating glutamate metabolism. YTHDF2 targets DEGS2 to regulate CRC lipid metabolism and inhibit CRC progression. The role of m6A “erasers” in the metabolic microenvironment in which CRC resides is still unclear
Hypoxic microenvironment
The hypoxic microenvironment is an important component of the TME and is closely associated with the malignant progression and poor prognosis of CRC (Rainho et al. 2021). The hypoxic microenvironment directly induces proliferation, invasion, metabolism, and genetic instability in CRC (Ulivi et al. 2016). In addition, hypoxia plays a crucial role in driving tumor angiogenesis, and leading to a vicious cycle of hypoxia and angiogenesis within tumors (Ulivi et al. 2016). Previous studies have demonstrated that changes in post-transcriptional m6A modifications significantly influence the hypoxic response (Xu et al. 2020a). Recent evidence also suggests that the hypoxic microenvironment acts as novel epigenetic mechanism that promotes CRC metastasis and is closely associated with m6A regulators.
Ruan et al. found that the m6A demethylase FTO was downregulated in CRC, and elevated levels of FTO were associated with a better prognosis for CRC patients. FTO downregulated the transcription of m6A downstream target gene MTA1, which inhibits CRC cell growth and metastasis in vivo. However, the context of hypoxic microenvironment decreased the FTO expression and weakened its regulatory activity, which was found to be HIF-1α independent (Ruan et al. 2021). The specific mechanism by which the hypoxic microenvironment downregulates FTO expression remains unclear. Yang et al. reported a significant co-expression of METTL3 and HIF-1α, which may be due to METTL3 regulating the translation efficiency of HIF-1α. The expression of METTL3 and total levels of m6A were significantly increased in CRC cell lines under hypoxic conditions. METTL3 knockdown inhibited CRC progression under hypoxic conditions. In addition, the m6A recognition factor YTHDF1 plays a crucial role as an m6A reader by binding to the m6A motif, regulating mRNA translation. YTHDF1 was significantly enriched in HIF-1α mRNA under hypoxic microenvironment. The increased expression of YTHDF1 promotes the formation of the hypoxic microenvironment by enhancing HIF-1α expression (Yang et al. 2021). Based on current studies, the hypoxic microenvironment interacts with the biological activity of CRC by regulating m6A modifications, and intervention of m6A regulators can in turn affect the progression of CRC by regulating the hypoxic microenvironment. Given the close relationship between the hypoxic microenvironment and CRC proliferation and metastasis, m6A regulators may be promising predictors and therapeutic targets for CRC prognosis. Figure 3 summarizes the role of m6A regulators in the CRC hypoxic microenvironment.
m6A modification interacts with the hypoxic microenvironment of CRC. METTL3 mediates the progression of EMT by enhancing the expression of HIF-1α. YTHDF1 is induced by the hypoxic microenvironment and promotes CRC progression by targeting HIF-1α. FTO is inhibited by the hypoxic microenvironment and suppresses CRC metastasis by suppressing the expression of MTA1
Inflammatory microenvironment
Chronic inflammation plays a key role in tumor progression, as demonstrated by various epidemiological and experimental studies. The inflammatory microenvironment has been closely associated with the development of CRC. Systemic inflammation is a poor prognosis marker in approximately 20–40% of CRC patients (Park et al. 2017). Inflammatory bowel disease (IBD) is also an independent risk factor for CRC, with inflammatory cells, cytokines and their associated inflammatory signaling pathways contributing to the establishment of an intestinal inflammatory microenvironment (Zhang and Qiao 2022). Increased m6A methylation has been demonstrated in major inflammatory pathways, including IL-6, TNF, and NF-κB signaling pathways. Additionally, m6A regulators are also involved in regulating inflammatory response of the tumor cells (Hou et al. 2019), (Chokkalla et al. 2019).
Bioinformatic analyses have revealed extensive interactions between m6A regulators and IBD risk genes. Moreover, m6A-related genes are significantly altered in IBD, and the IBD risk locus is also modified by m6A (Nie et al. 2021; Sebastian-delaCruz et al. 2020). Those findings suggested that m6A modifications regulated intestinal inflammatory environment are closely associated with the development of IBD. The persistent inflammatory microenvironment may increase the expression of m6A and m6A regulatory proteins. Cluster analysis based on m6A features revealed that subgroups of m6AregC1 (characterized with high expression of IGF2BP3) and m6AsigC1 (characterized with immune activation, with high CD8+ effector T cells, transcripts of immune activation, and immune checkpoints) were characterized by activation of inflammatory pathways and infiltration of inflammatory cells, and these subgroups were more responsive for CRC immunotherapy (Zhang et al. 2022b). Therefore, assessing m6A levels in CRC could provide insights into the inflammatory microenvironment and help in modulating it for better immunotherapy outcomes. Furthermore, the m6A methyltransferase METTL3 has been found to promote the formation of an inflammatory microenvironment and CRC cell proliferation by inhibiting SOCS2 (Li et al. 2021b; Xu et al. 2020b). Additionally, m6A methylation of EphA2 and VEGFA regulated by IGF2BP2/3, induces an inflammatory microenvironment and promotes angiogenesis in CRC via PI3K/AKT and ERK1/2 signaling pathway (Liu et al. 2022a). Macrophages play a crucial role in the inflammatory response, with two polarized subtypes, M1 and M2. M1 macrophages exhibit pro-inflammatory and anti-tumor phenotypes, while M2 macrophages exert anti-inflammatory effects and are involved in tumor metastasis (Li et al. 2018). Elevated activity of METTL3 has been found to directly methylate and stabilize STAT1 mRNA, leading to M1 macrophage polarization and inhibition of M2 macrophage formation (Liu et al. 2019).
Together, m6A modification regulates the formation of the inflammatory microenvironment, while the inflammatory microenvironment also influences m6A modification. However, the most current studies on m6A modifications and the inflammatory microenvironment of CRC are based on bioinformatics analysis and require further validation in vitro and in vivo studies. Figure 4 provides and overview of the role of m6A regulators in the CRC inflammatory microenvironment.
m6A modifications interact with the CRC inflammatory microenvironment. METTL3 promotes the formation of the inflammatory microenvironment and CRC proliferation by inhibiting SOCS2. Additionally, METTL3 promotes M1-type macrophage polarization and inhibits M2-type macrophage by promoting STAT1 expression. The m6A methylated EphA2/VEGFA promotes angiogenesis in CRC by targeting the PI3K/AKT inflammatory signaling pathway. The functions of m6A “erasers” in the inflammatory microenvironment of CRC are still unclear
Immune microenvironment
TME alters the proliferation, metastasis, and prognosis of CRC. For instance, it induces immune tolerance and immunosuppression, enabling CRC cells to evade the immune system (Shen et al. 2020). The immune components of TME present attractive targets for cancer therapy across various types of cancer. Emerging studies indicated that m6A methylation plays a crucial role in regulating the immune microenvironment in CRC.
T cells play a crucial role in regulating the entire adaptive immune response. Recent studies have shown that m6A can influence the selective differentiation of tumor-infiltrating T cells by targeting various protein components or signaling pathways (Li et al. 2021c). For instance, increased m6A methylation mediated by xeroderma pigmentosum complementation group G (XPG) leads to the release of IFN-γ from Th1 cells. This in turn induces CTL activity and activates CD8+CTL (Pal et al. 2022). Additionally, m6A targets the SOCS protein family, regulates the IL-7 and TCR signaling pathways, and influences the direction of homeostatic proliferation and differentiation of naive T cells (Li et al. 2017b). Dong et al. found that decreased levels of overall m6A and METTL14 in CRC tumor stromal cells were associated with reduced T cell infiltration in CRC patients. Interestingly, this study also revealed that repressive macrophages from CRC patients interacted with CD8+ T cells in TME (Dong et al. 2021). The immunosuppressive TME inhibits the cytotoxic T cells function and promotes T cell exhaustion, ultimately leading to tumor evasion. Another clinical study has demonstrated that METTL14 deficiency is associated with a poor prognosis in CRC patients (Yang et al. 2020). LncRNA XIST, a target of METTL14, is closely associated with T-cell immunity. XIST regulates the immune function of CD8+T cells through the miR-34a-5p/PDL1 axis and promotes Th17 differentiation through the KDM6A-TSAd pathway (Li et al. 2022b; Syrett et al. 2019). Deletion of METTL14 significantly reduces the m6A level of XIST, leading to increased XIST expression and enhanced proliferative and invasive abilities of CRC cells (Yang et al. 2020). However, conflicting results have also been reported. Wang et al. found that deficiency of METTL3/14 in CRC cells stabilizes SATA1 through YTHDF2. This stabilization leads to increased IFN-γ secretion and CD8+T cell infiltration (Wang et al. 2020b). Myeloid-derived suppressor cells (MDSCs) are known for their strong immunosuppressive activities, and promotes the formation of immunosuppressive microenvironment. Chen et al. found that METTL3 promotes the expression of BHLHE41 in an m6A-dependent manner, which induces CXCL1 transcription and enhances the migration of MDSCs in vitro. Inhibition of METTL3 expression in CRC cells reduces the MDSCs accumulation, maintains the activation and proliferation of CD4+ T cells and CD8+ T cells, and inhibits the progression of CRC (Chen et al. 2022). Therefore, targeting MDSCs by m6A methylation modifications could be a promising strategy for anti-cancer therapy. The inconsistencies observed in these studies may be caused by differences in the downstream targets and modification sites of METTL3 and METTL14. However, the alteration of m6A affects the immune microenvironment of CRC, confirming the critical role of m6A "readers" in tumor immune surveillance. DCs are responsible for antigen processing, presentation and activation of the T-cell immune response. Meanwhile, a large number of aberrant m6A modifications have been found in DCs of tumors (Shulman and Stern-Ginossar 2020). Deletion of YTHDF1 in DCs enhances cross-presentation of CRC antigens and activates CD8+T cells in vivo. YTHDF1-deficient CRC mice also exhibit higher sensitivity to immunotherapy (Han et al. 2019a). In addition, CD34 and CD276 have been reported as molecular predictors for the viability of CRC patients, reshaping the immune microenvironment of CRC in an m6A-dependent manner, and mediating the immune escape mechanism of CRC by regulating immune checkpoints such as CTLA-4 (Zhou et al. 2021).
Together, m6A regulators play crucial roles in the formation of the diversity and complexity of the immune microenvironment, and regulate immunosuppression and/or immune escape. To further unravel the relationship between m6A modification and immune microenvironment, it is necessary to investigate the effects of m6A methylation on the functional and biological behavior of immune cells (e.g., metabolism), as well as the mechanisms of cross-talk between tumor cells, immune cells, and additional stromal cells. The role of m6A including the m6A-associated immune regulatory network, also requires further investigation in the immune microenvironment of CRC. Figure 5 illustrates the role of m6A regulators in the immune microenvironment of CRC.
m6A modifications interact with the immune microenvironment of CRC. In m6A “writers”, METTL3 promotes infiltration of MDSCs and suppresses CD8+ T cells by targeting BHLHE41. METTL3 promotes the stem cell-like phenotype of CRC by targeting Sec62. METTL3 promotes CRC progression by targeting NCALD and TCF7L2. METTL14 represses the proliferation of CRC by targeting XIST expression. In m6A “Readers”, YTHDF1 inhibits DCs antigen presentation and CD8+T cells activation. YTHDF3 promotes the translation of drug-resistance genes by recruiting eLF2AK2. In m6A “Erasers”, inhibition of FTO deregulates PD-L1 expression and CRC progression. ALKBH5 inhibits the infiltration of Tregs and MDSCs and the progression of CRC by targeting Mct4/Slc16a3
Therapeutic strategies for CRC based on m6A modification
Immunotherapy
Immunotherapy is a promising cancer treatment that assists the immune system against tumor cells. The current immunotherapy strategies include monoclonal antibodies, lysing viruses, tumor vaccines, immune checkpoint inhibitors (ICIs), and adoptive transfer. Particularly, immune checkpoint inhibitors have shown remarkable effects. Additionally, the m6A modification involved in ICI immunotherapy provides alternative approaches for CRC treatment (Shriwas et al. 2020), (Lichtenstern et al. 2021).
Inhibition of METTL3/14 improves the sensitivity of CRC to anti-PD-1 treatment by impairing m6A modification, thereby altering the TME and CD8+ T cells recruitment (Wang et al. 2020b). Li et al. discovered that m6A demethylase ALKBH5-deficient CRC mice exhibited significantly elevated viability after anti-PD-1 treatment. Although ALKBH5 is not required for CRC growth and survival in vivo or in vitro, it plays a critical role in the effectiveness of anti-PD-1 therapy (Li et al. 2020b). The expression of m6A recognition protein YTHDF1 correlates with the outcome of immunotherapy in CRC patients. YTHDF1 deletion strengthens the anti-tumor effect of PD-1 blockers by restoring the infiltration of CD8+T cells (Li et al. 2023b). The m6A demethylase FTO is associated with the progression of various tumors. FTO deletion inhibits PD-L1 expression in CRC cells, and this process is independent of IFN-γ signaling (Tsuruta et al. 2020). Furthermore, numerous bioinformatics analyses have revealed that m6A modifications are involved in shaping tumor immune microenvironment profiles (Chen et al. 2021a), (Liu et al. 2022b). All the findings highlight the significant role of m6A modifications in modulating the responsiveness of CRC to immunotherapy, and the m6A modulators may serve as potential therapeutic targets for CRC alone or in combination with immune checkpoint inhibitors.
Chemotherapy
Drug resistance is the primary cause of failure in cancer chemotherapy. The mechanisms underlying tumor drug resistance include in drug metabolism, tumor heterogeneity, microenvironmental alterations, and mutations in target proteins (Vasan et al. 2019).
Oxaliplatin (OX) is widely used first-line chemotherapeutic agent for cancer treatment, but resistance developed by tumor cells poses a major challenge in the treatment of advanced CRC. Lan et al. discovered that the total m6A content and the expression of methyltransferase METTL3 increased in CRC tissues from OX-resistant patients. TAM in TME contribute to OX-resistance in CRC cells via METTL3-mediated m6A modification (Lan et al. 2021). METTL3-induced m6A modification increases the stability of Sec62 mRNA and upregulates Sec62 expression, which maintains the stem cell-like phenotype and chemotherapy resistance in CRC (Liu et al. 2021). IR100HG stabilizes TCF7L2 mRNA with METTL3-mediated m6A modification to regulate CRC resistance to cetuximab. TCF7L2 in turn regulates the transcription of MIR100HG and blocks the positive feedback pathway between them (Gao et al. 2021; Liu et al. 2022c). METTL3 is associated with 5-fluorouracil (5-FU) resistance in CRC. METTL3-mediated m6A modification acts on DiGeorge syndrome critical region 8 (DGCR8) in CAFs, promoting the secretion of exosomal miR-181b-5p, which inhibits CRC sensitivity to 5-FU by targeting NCALD (Pan et al. 2022). Moreover, the YTHDF family has also been implicated in drug resistance in CRC. Expression of YTHDF1 is significantly upregulated in cisplatin-resistant CRC cell lines and promotes CRC resistance to cisplatin by mediating glutamine metabolism (Chen et al. 2021b). Moreover, YTHDF1 is targeted by miR-136-5p to mediate CRC chemoresistance (Jiang et al. 2022). YTHDF3 is highly expressed in OX-resistant CRC tissues and recognizes the 5′-UTR of m6A-methylated RNAs associated with tumor resistance, and recruiting eukaryotic translation initiation factor 3 subunit A (eIF3A) to promote translation of drug-resistant genes (Zhao et al. 2022). All the studies demonstrate that m6A modification mediates chemoresistance in CRC by altering various cell types in the TME. Consequently, targeting m6A regulators may offer a new horizon for addressing chemoresistance of CRC.
Discussion
TME plays a critical role in the development of human cancers (Hinshaw and Shevde 2019). The prevalence and aberrant distribution of m6A modifications are involved in tumor development (Li et al. 2021d; Han et al. 2019b). The “writers”, “erasers”, and “readers” of m6A dynamically regulate the components of the TME through complex pathways. These pathways influence the metabolic, hypoxic, inflammatory, and immune microenvironment (Fang et al. 2022). As m6A regulators affect multiple tumorous microenvironments, blocking one pathway may result in compensatory expression in another pathway. This highlights the need for co-blockade of multiple pathways to achieve synergistic anti-tumor activity. Table 1 provides a summary of the effects and targets of m6A regulators in the CRC microenvironment.
In addition, m6A modifications enable cross-talk with different microenvironments. Metabolites produced by gut microbes regulate the levels of m6A in different cell types, consequently affecting cellular activity in TME. Furthermore, alterations in metabolism, induced by the metabolic microenvironment and hypoxic condition, contribute to the development of a chronic inflammatory microenvironment. This, in turn, suppresses immune function within the gut and provides favorable conditions for progression of CRC. The m6A regulatory factor exhibits diverse effects in different microenvironments, and its response functions can be considered a double-edged sword in CRC. For instance, METTL3 has been proposed as a tumor-promoting factor, whereas it also plays a role in polarizing M1 macrophages and exhibits anti-tumor effects (Gunassekaran et al. 2021).
Exosomes facilitate the transfer and exchange of miRNAs, mRNAs, and lncRNAs between cells and tissues, and have been suggested to play a critical role in regulating the TME (Wortzel et al. 2019). Interestingly, recent studies have proposed that the inter-regulatory relationship between exosomes and m6A is associated with tumor tolerance to chemotherapy and radiotherapy, possibly by influencing the TME (Song et al. 2021). Additionally, the key to successful therapy targeting the m6A enzyme lies in safely and efficiently delivering the therapeutic agent to specific cells. Firstly, these carriers must protect the cargo from destruction, and secondly, they must bind to specific cells and enter them to release the cargo. This requires an effective drug delivery route and specific targeting molecules on the carrier surface to attract receptors on the target cell surface. Taking these conditions into account, viral delivery systems, lipid nanoparticle delivery systems, and virus-like particle delivery systems show promise for targeting m6A enzymes in specific cells (Raguram et al. 2022). Currently available techniques for measuring m6A activity include high-throughput sequencing, colorimetry, and liquid chromatography-mass spectrometry, such as MeRIP-seq, miCLIP-seq, SCARLET, and LC–MS/MS. Using these techniques to measure the m6A activity of specific genes in CRC cells holds potential as a diagnostic tool for monitoring the progress of CRC.
Although the emergence of m6A regulators has provided new ideas for CRC treatment, there are still numerous challenges regarding the regulatory role of m6A modifications on TME and related applications. Firstly, m6A modifications are abundant in TME, but most studies have not detected specific biological functions of the associated regulators, limiting further exploration of their applications. Secondly, since RNA status varies across individuals, tissues, and cell types, it is difficult to target m6A modifications to specific cell types in different individuals. Additionally, m6A modification is a complex regulatory network, and the current understanding of the relevant regulatory factors of m6A is incomplete, leaving more m6A “writers”, “erasers” and “readers” to be uncovered. Finally, few TME-related studies in CRC currently involve the process of m6A modification in vivo, with most being bioinformatics analyses or in vitro assays. In contrast, m6A modification is a dynamic regulatory process in vivo, posing a major obstacle to applying relevant findings to clinical practice.
Conclusion
The TME is a complex and dynamic system that encompasses hundreds of chemical modifications. These modifications serve to activate and influence signaling mechanisms for epigenetic changes. Among these modifications, the m6A modifications play a crucial role in regulating the dynamics of the TME and have a profound impact on the metabolic, hypoxic, inflammatory, and immune microenvironment of CRC. The m6A modulators have extensive applications in settings and show great potential as novel biomarkers or targets for interventions in CRC. Therefore, it is essential to further comprehend the regulatory mechanisms of m6A modifications in the TME in order to explore the oncogenes and biological behaviors associated with CRC.
Availability of data and materials
Not applicable.
Abbreviations
- 5-FU:
-
5-Fluorouracil
- ALKBH3:
-
α-Ketoglutarate-dependent dioxygenase homolog 3
- ALKBH5:
-
α-Ketoglutarate-dependent dioxygenase homolog 5
- CAFs:
-
Cancer-associated fibroblasts
- CBLL1:
-
Cbl proto-oncogene, E3 ubiquitin protein ligase-like 1
- CCNE1:
-
Cell cycle protein E1
- CRC:
-
Colorectal cancer
- DCs:
-
Dendritic cells
- DGCR8:
-
DiGeorge syndrome critical region 8
- eLF2AK2:
-
Eukaryotic translation initiation factor 3 subunit A
- ETBF:
-
Enterotoxigenic Bacteroides Fragilis
- FTO:
-
Fat mass and obesity-associated protein
- HIF-1α:
-
Hypoxia-inducible factor 1-α
- HNRNPA2B1:
-
Heterogeneous nuclear ribonucleoprotein A2B1
- HNRNPC:
-
Heterogeneous nuclear ribonucleoprotein C
- IBD:
-
Inflammatory bowel disease
- IGF2BPs:
-
Insulin-like growth factor 2 mRNA-binding proteins
- IMPs:
-
IGF-II mRNA-binding proteins
- m6A:
-
N6-methyladenosine
- MDSCs:
-
Myeloid-derived suppressor cells
- METTL14:
-
Methyltransferase-like 14
- METTL16:
-
Methyltransferase-like 16
- METTL3:
-
Methyltransferase-like 3
- ncRNAs:
-
Non-coding RNAs
- OX:
-
Oxaliplatin
- RBM15:
-
RNA-binding motif protein 15
- RBM15B:
-
RNA-binding motif protein 15B
- TAMs:
-
Tumor-associated macrophages
- TANs:
-
Tumor-associated neutrophils
- TME:
-
Tumor microenvironment
- UC:
-
Ulcerative colitis
- WTAP:
-
Wilms’ tumor 1-associated protein
- YTHDC:
-
YT521-B homology domain containing proteins
- YTHDF:
-
YT521-B homology domain family proteins
- ZC3H13:
-
Zinc finger CCCH-type containing 13
References
Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015a;519:482–5.
Alarcón C, et al. HNRNPA2B1 is a mediator of m (6)A-dependent nuclear RNA processing events. Cell. 2015b; S0092867415010247.
Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500.
An Y, Duan H. The role of m6A RNA methylation in cancer metabolism. Mol Cancer. 2022;21:14.
Arneth B. Tumor microenvironment. Medicina (kaunas). 2019;56:15.
Arnold M, et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–91.
Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–47.
Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (engl). 2021a;134:783–91.
Cao Y, et al. Enterotoxigenic bacteroidesfragilis promotes intestinal inflammation and malignancy by inhibiting exosome-packaged miR-149-3p. Gastroenterology. 2021b;161(1552–1566): e1512.
Catana CS, et al. Contribution of the IL-17/IL-23 axis to the pathogenesis of inflammatory bowel disease. World J Gastroenterol. 2015;21:5823–30.
Chen Z, et al. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 2019;47:2533–45.
Chen H, et al. Cross-talk of four types of RNA modification writers defines tumor microenvironment and pharmacogenomic landscape in colorectal cancer. Mol Cancer. 2021a;20:29.
Chen P, et al. Targeting YTHDF1 effectively re-sensitizes cisplatin-resistant colon cancer cells by modulating GLS-mediated glutamine metabolism. Mol Ther Oncolytics. 2021b;20:228–39.
Chen H, et al. METTL3 inhibits antitumor immunity by targeting m (6)A-BHLHE41-CXCL1/CXCR2 axis to promote colorectal cancer. Gastroenterology. 2022;163:891–907.
Chokkalla AK, et al. Transient focal ischemia significantly alters the m (6)A epitranscriptomic tagging of RNAs in the brain. Stroke. 2019;50:2912–21.
Dai D, Wang H, Zhu L, Jin H, Wang X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018;9:124.
Deng X, et al. RNA N (6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28:507–17.
Dong L, et al. The loss of RNA N (6)-adenosine methyltransferase Mettl14 in tumor-associated macrophages promotes CD8 (+) T cell dysfunction and tumor growth. Cancer Cell. 2021;39(945–957): e910.
Fang Z, et al. Role of m6A writers, erasers and readers in cancer. Exp Hematol Oncol. 2022;11:45.
Feingold M, Hall BD, Lacassie Y, Martinez-Frias ML. Syndrome of microcephaly, facial and hand abnormalities, tracheoesophageal fistula, duodenal atresia, and developmental delay. Am J Med Genet. 1997;69:245–9.
Gao Q, et al. A WNT7B-m (6)A-TCF7L2 positive feedback loop promotes gastric cancer progression and metastasis. Signal Transduct Target Ther. 2021;6:43.
Gunassekaran GR, Poongkavithai Vadevoo SM, Baek MC, Lee B. M1 macrophage exosomes engineered to foster M1 polarization and target the IL-4 receptor inhibit tumor growth by reprogramming tumor-associated macrophages into M1-like macrophages. Biomaterials. 2021;278: 121137.
Guo W, et al. M6A methylation of DEGS2, a key ceramide-synthesizing enzyme, is involved in colorectal cancer progression through ceramide synthesis. Oncogene. 2021;40:5913–24.
Han D, et al. Anti-tumour immunity controlled through mRNA m (6)A methylation and YTHDF1 in dendritic cells. Nature. 2019a;566:270–4.
Han J, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019b;18:110.
Han Z, et al. RNA m (6)A modification in prostate cancer: A new weapon for its diagnosis and therapy. Biochim Biophys Acta Rev Cancer. 2023;1878: 188961.
He L, et al. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18:176.
Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79:4557–66.
Hou J, et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol Cancer. 2019;18:163.
Huang H, et al. Recognition of RNA N (6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–95.
Huang J, Shao Y, Gu W. Function and clinical significance of N6-methyladenosine in digestive system tumours. Exp Hematol Oncol. 2021;10:40.
Jackson DN, Theiss AL. Gut bacteria signaling to mitochondria in intestinal inflammation and cancer. Gut Microbes. 2020;11:285–304.
Jia G, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–7.
Jiang X, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6:74.
Jiang Z, et al. Circular RNA protein tyrosine kinase 2 (circPTK2) promotes colorectal cancer proliferation, migration, invasion and chemoresistance. Bioengineered. 2022;13:810–23.
Jordan KR, Loman BR, Bailey MT, Pyter LM. Gut microbiota-immune-brain interactions in chemotherapy-associated behavioral comorbidities. Cancer. 2018;124:3990–9.
Lan H, et al. Tumor-associated macrophages promote oxaliplatin resistance via METTL3-mediated m (6)A of TRAF5 and necroptosis in colorectal cancer. Mol Pharm. 2021;18:1026–37.
Li A, et al. Cytoplasmic m (6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017a;27:444–7.
Li HB, et al. m (6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017b;548:338–42.
Li C, et al. Macrophage polarization and meta-inflammation. Transl Res. 2018;191:29–44.
Li T, et al. METTL3 facilitates tumor progression via an m (6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18:112.
Li K, Luo H, Luo H, Zhu X. Clinical and prognostic pan-cancer analysis of m6A RNA methylation regulators in four types of endocrine system tumors. Aging-Us. 2020a;12:23931–44.
Li N, et al. ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc Natl Acad Sci U S A. 2020b;117:20159–70.
Li Q, et al. HIF-1alpha-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. Signal Transduct Target Ther. 2021a;6:76.
Li S, et al. SOCS2 suppresses inflammation and apoptosis during NASH progression through limiting NF-kappaB activation in macrophages. Int J Biol Sci. 2021b;17:4165–75.
Li M, Zha X, Wang S. The role of N6-methyladenosine mRNA in the tumor microenvironment. Biochim Biophys Acta Rev Cancer. 2021c;1875: 188522.
Li Y, et al. Molecular characterization, biological function, tumor microenvironment association and clinical significance of m6A regulators in lung adenocarcinoma. Brief Bioinform 2021d;22.
Li Y, He L, Wang Y, Tan Y, Zhang F. N (6)-methyladenosine methyltransferase KIAA1429 elevates colorectal cancer aerobic glycolysis via HK2-dependent manner. Bioengineered. 2022a;13:11923–32.
Li J, et al. XIST/miR-34a-5p/PDL1 axis regulated the development of lung cancer cells and the immune function of CD8 (+) T cells. J Recept Signal Transduct Res. 2022b;42:469–78.
Li J, et al. Promotive role of USP29-mediated deubiquitination in malignant proliferation of colorectal cancer cells via the KIAA1429/SOX8 axis. Biomol Biomed. 2023a;23:483–95.
Li T, et al. Methionine deficiency facilitates antitumour immunity by altering m (6)A methylation of immune checkpoint transcripts. Gut. 2023b;72:501–11.
Lichtenstern CR, Ngu RK, Shalapour S, Karin M. Immunotherapy, inflammation and colorectal cancer. Cells. 2020; 9.
Liu J, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–5.
Liu Y, et al. The N (6)-methyladenosine (m (6)A)-forming enzyme METTL3 facilitates M1 macrophage polarization through the methylation of STAT1 mRNA. Am J Physiol Cell Physiol. 2019;317:C762–75.
Liu X, et al. Sec62 promotes stemness and chemoresistance of human colorectal cancer through activating Wnt/beta-catenin pathway. J Exp Clin Cancer Res. 2021;40:132.
Liu X, et al. m6A methylated EphA2 and VEGFA through IGF2BP2/3 regulation promotes vasculogenic mimicry in colorectal cancer via PI3K/AKT and ERK1/2 signaling. Cell Death Dis. 2022a;13:483.
Liu T, et al. Comprehensive analysis of m (6)A regulator-based methylation modification patterns characterized by distinct immune profiles in colon adenocarcinomas. Gene. 2022b;821: 146250.
Liu H, et al. Interaction of lncRNA MIR100HG with hnRNPA2B1 facilitates m (6)A-dependent stabilization of TCF7L2 mRNA and colorectal cancer progression. Mol Cancer. 2022c;21:74.
Liu Y, et al. N6-methyladenosine-mediated gene regulation and therapeutic implications. Trends Mol Med. 2023;29:454–67.
Lma B, et al. The m6A reader YTHDC2 inhibits lung adenocarcinoma tumorigenesis by suppressing SLC7A11-dependent antioxidant function—ScienceDirect. Redox Biol. 2020;38:101801.
Lu S, et al. N6-methyladenosine reader IMP2 stabilizes the ZFAS1/OLA1 axis and activates the Warburg effect: implication in colorectal cancer. J Hematol Oncol. 2021;14:188.
Lv W, et al. Analysis and validation of m6A regulatory network: a novel circBACH2/has-miR-944/HNRNPC axis in breast cancer progression. J Transl Med. 2021;19:527.
Marcinkowski M, et al. Human and Arabidopsis alpha-ketoglutarate-dependent dioxygenase homolog proteins-New players in important regulatory processes. IUBMB Life. 2020;72:1126–44.
Maybin JA, et al. Hypoxia and hypoxia inducible factor-1alpha are required for normal endometrial repair during menstruation. Nat Commun. 2018;9:295.
Nie K, et al. A broad m6A modification landscape in inflammatory bowel disease. Front Cell Dev Biol. 2021;9: 782636.
Ocvirk S, O’Keefe SJD. Dietary fat, bile acid metabolism and colorectal cancer. Semin Cancer Biol. 2021;73:347–55.
Pal R, et al. Involvement of xeroderma pigmentosum complementation group G (XPG) in epigenetic regulation of T-Helper (T (H)) cell differentiation during breast cancer. Immunobiology. 2022;227: 152259.
Pan S, et al. N6‑methyladenosine upregulates miR‑181d‑5p in exosomes derived from cancer‑associated fibroblasts to inhibit 5‑FU sensitivity by targeting NCALD in colorectal cancer. Int J Oncol. 2022; 60.
Park JH, et al. Tumour invasiveness, the local and systemic environment and the basis of staging systems in colorectal cancer. Br J Cancer. 2017;116:1444–50.
Raguram A, Banskota S, Liu DR. Therapeutic in vivo delivery of gene editing agents. Cell. 2022;185:2806–27.
Rainho MA, Mencalha AL, Thole AA. Hypoxia effects on cancer stem cell phenotype in colorectal cancer: a mini-review. Mol Biol Rep. 2021;48:7527–35.
Roca-Lema D, et al. Role of the E3 ubiquitin-ligase Hakai in intestinal inflammation and cancer bowel disease. Sci Rep. 2022;12:17571.
Ruan DY, et al. FTO downregulation mediated by hypoxia facilitates colorectal cancer metastasis. Oncogene. 2021;40:5168–81.
Ruszkowska A. METTL16, methyltransferase-like protein 16: current insights into structure and function. Int J Mol Sci. 2021;22:2176.
Scholler E, et al. Interactions, localization, and phosphorylation of the m (6)A generating METTL3-METTL14-WTAP complex. RNA. 2018;24:499–512.
Sebastian-delaCruz M, et al. Implication of m6A mRNA methylation in susceptibility to inflammatory bowel disease. Epigenomes. 2020;4:16.
Shen F, et al. Oxaliplatin-/NLG919 prodrugs-constructed liposomes for effective chemo-immunotherapy of colorectal cancer. Biomaterials. 2020;255: 120190.
Shriwas O, Mohapatra P, Mohanty S, Dash R. The impact of m6A RNA modification in therapy resistance of cancer: implication in chemotherapy, radiotherapy, and immunotherapy. Front Oncol. 2020;10: 612337.
Shulman Z, Stern-Ginossar N. The RNA modification N (6)-methyladenosine as a novel regulator of the immune system. Nat Immunol. 2020;21:501–12.
Slavin JL. Position of the American Dietetic Association: health implications of dietary fiber. J Am Diet Assoc. 2008;108:1716–31.
Song Z, Jia G, Ma P, Cang S. Exosomal miR-4443 promotes cisplatin resistance in non-small cell lung carcinoma by regulating FSP1 m6A modification-mediated ferroptosis. Life Sci. 2021;276: 119399.
Sung H, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49.
Syrett CM, et al. Altered X-chromosome inactivation in T cells may promote sex-biased autoimmune diseases. JCI Insight. 2019;4.
Tsuruta N, et al. RNA N6-methyladenosine demethylase FTO regulates PD-L1 expression in colon cancer cells. Biochem Biophys Res Commun. 2020;530:235–9.
Ulivi P, Marisi G, Passardi A. Relationship between hypoxia and response to antiangiogenic therapy in metastatic colorectal cancer. Oncotarget. 2016;7:46678–91.
Vasan N, Baselga J, Hyman DM. A view on drug resistance in cancer. Nature. 2019;575:299–309.
Wang P, Doxtader KA, Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell. 2016;63:306–17.
Wang Q, et al. METTL3-mediated m (6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020a;69:1193–205.
Wang L, et al. m (6) A RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy. EMBO J. 2020b;39: e104514.
Wortzel I, Dror S, Kenific CM, Lyden D. Exosome-mediated metastasis: communication from a distance. Dev Cell. 2019;49:347–60.
Wu T, Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017;387:61–8.
Xiao W, Zhao BS, Roundtree IA, Lu Z, He C. N (6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–99.
Xiao W, et al. Nuclear m (6)A reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61:507–19.
Xu K, et al. N (6)-methyladenosine demethylases Alkbh5/Fto regulate cerebral ischemia-reperfusion injury. Ther Adv Chronic Dis. 2020a;11:2040622320916024.
Xu J, et al. m6A methyltransferase METTL3 maintains colon cancer tumorigenicity by suppressing SOCS2 to promote cell proliferation. Oncol Rep. 2020b;44:973–86.
Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m (6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–24.
Yang X, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19:46.
Yang Z, et al. Knockdown of RNA N6-methyladenosine methyltransferase METTL3 represses Warburg effect in colorectal cancer via regulating HIF-1alpha. Signal Transduct Target Ther. 2021;6:89.
Yang J, et al. High-fat diet promotes colorectal tumorigenesis through modulating gut microbiota and metabolites. Gastroenterology. 2022;162(135–149): e132.
Zhang F, Qiao S. Research progress on the relationship between inflammation and colorectal cancer. Ann Gastroenterol Surg. 2022;6:204–11.
Zhang B, Gu Y, Jiang G. Expression and prognostic characteristics of m (6) A RNA methylation regulators in breast cancer. Front Genet. 2020;11: 604597.
Zhang K, et al. N (6)-methyladenosine-mediated LDHA induction potentiates chemoresistance of colorectal cancer cells through metabolic reprogramming. Theranostics. 2022a;12:4802–17.
Zhang Y, et al. Links between N (6)-methyladenosine and tumor microenvironments in colorectal cancer. Front Cell Dev Biol. 2022b;10: 807129.
Zhao Y, et al. YTHDF3 facilitates eIF2AK2 and eIF3A recruitment on mRNAs to regulate translational processes in oxaliplatin-resistant colorectal cancer. ACS Chem Biol. 2022;17:1778–88.
Zheng G, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29.
Zheng Y, et al. N6-methyladenosine modification of PTTG3P contributes to colorectal cancer proliferation via YAP1. Front Oncol. 2021;11: 669731.
Zhou Y, et al. Decreased m6A modification of CD34/CD276 (B7–H3) leads to immune escape in colon cancer. Front Cell Dev Biol. 2021;9: 715674.
Zhu D, et al. ZC3H13 suppresses colorectal cancer proliferation and invasion via inactivating Ras-ERK signaling. J Cell Physiol. 2019;234:8899–907.
Zhu W, et al. Methyltransferase like 3 promotes colorectal cancer proliferation by stabilizing CCNE1 mRNA in an m6A-dependent manner. J Cell Mol Med. 2020;24:3521–33.
Acknowledgements
Not applicable
Funding
This study was supported by the National Natural Science Foundation of China (82102901), the Innovation and Entrepreneurship Training Program for College Students by Yangzhou University (X20220742), and the Policy Guidance Program of Yangzhou (YZ2022234).
Author information
Authors and Affiliations
Contributions
JY drafted the manuscript in detail, drew and corrected figures, counted and plotted the table. YS researched literatures and supplementary the article content. XY supplementary the article content. ZL drafted the manuscript, drew and corrected figures, critically revised the article for important intellectual content. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yao, J., Song, Y., Yu, X. et al. Interaction between N6-methyladenosine modification and the tumor microenvironment in colorectal cancer. Mol Med 29, 129 (2023). https://doi.org/10.1186/s10020-023-00726-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s10020-023-00726-2