Abstract
Background
The quantitative structure property relationship (QSPR) for octanol/air partition coefficient (KOA) of polybrominated diphenyl ethers (PBDEs) was investigated. Molecular distance-edge vector (MDEV) index was used as the structural descriptor of PBDEs. The quantitative relationship between the MDEV index and the lgKOA of PBDEs was modeled by multivariate linear regression (MLR) and artificial neural network (ANN) respectively. Leave one out cross validation and external validation was carried out to assess the predictive ability of the developed models. The investigated 22 PBDEs were randomly split into two groups: Group I, which comprises 16 PBDEs, and Group II, which comprises 6 PBDEs.
Results
The MLR model and the ANN model for predicting the KOA of PBDEs were established. For the MLR model, the prediction root mean square relative error (RMSRE) of leave one out cross validation and external validation is 2.82 and 2.95, respectively. For the L-ANN model, the prediction RMSRE of leave one out cross validation and external validation is 2.55 and 2.69, respectively.
Conclusion
The developed MLR and ANN model are practicable and easy-to-use for predicting the KOA of PBDEs. The MDEV index of PBDEs is shown to be quantitatively related to the KOA of PBDEs. MLR and ANN are both practicable for modeling the quantitative relationship between the MDEV index and the KOA of PBDEs. The prediction accuracy of the ANN model is slightly higher than that of the MLR model. The obtained ANN model shoud be a more promising model for studying the octanol/air partition behavior of PBDEs.

Similar content being viewed by others
Background
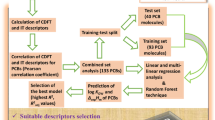
Polybrominated diphenyl ethers (PBDEs) are a series of organobromine compounds that have been widely used as flame retardant in a variety of products, such as building materials, electronics, furnishings, coatings, plastics, etc [1, 2]. Although the production of some PBDEs has been restricted under the Stockholm Convention since 2010, PBDEs have already become ubiquitous pollutants in the environment. They have been detected in many environmental compartments, such as air, water, soil, vegetations, animals and humans [3, 4]. PBDEs have gained increasing attention because of their environmental persistence, bioaccumulation through the food chain, and potential risk to the human health [1, 5, 6]. PBDEs are lipophilic and semi-volatile compounds. The octanol/air partition of PBDEs may influence their fate, transport, and transformation in atmospheres [7–9]. The octanol/air partition coefficient (KOA), which is defined as the ratio of solute concentration in air versus octanol when the octanol/air system is at equilibrium, is a key parameter for describing the octanol/air partition of PBDEs between the atmosphere and organic phases such as soil, aerosol, vegetation and animals. Thus, a quantitative study on the KOA of PBDEs is of great importance to understand the environmental fate of PBDEs. Many efforts have been made to determine the KOA of PBDEs [7, 9–11]. However, determining the KOA of PBDEs is always a hard work due to the complexity of analytical methods, lack of chemical standards and high cost of experiments [4, 12–15]. Thus, the quantitative structure-property relationship (QSPR) method, which is fast, easy-to-use and cost-effective [12, 16, 17], is always used to preliminary estimate the value of KOA of PBDEs. Several QSPR models for the KOA of PBDEs have been reported [12–15]. In these works, quantum chemical descriptors are used as the structural descriptor of PBDEs. However, developing a QSPR model based on quantum chemical descriptors is still a complex work, because the calculation and selection of structural descriptors are always time-consuming and complicated. It is still worthwhile to develop an easy-to-use QSPR model for the KOA of PBDEs. Topological index is a kind of structural descriptor which has been widely used in the QSPR researches. It can effectively describe the structure of molecules without the detailed molecular orbital calculation and energy optimization. Topological index is useful because, despite its mathematical simplicity, it is able to differentiate molecules with different structures [18]. Therefore, the aim of our work is to investigate the QSPR model for the KOA of PBDEs based on topological index. Molecular distance-edge vector (MDEV) index [19–21] was used as the structural descriptor of PBDEs. Multivariate linear regression (MLR) and artificial neural network (ANN) were employed to build the calibration model between the MDEV index and the KOA of PBDEs.
Results and discussion
Firstly, the MDEV index of the investigated 22 PBDEs was calculated. The obtained MDEV index is presented in Table 1. As shown in the table, the value of MDEV index for different PBDE molecules is different. It is demonstrated that MDEV index can describe the structural differences among these molecules. Thus, it is reasonable to use MDEV index as structural descriptor to develop the QSPR model of PBDEs.
Secondly, two QSPR models were developed and investigated. One is MLR model and the other is L-ANN model. In order to assess the predictive ability of the developed models, two validation methods, leave one out cross validation and external validation, were conducted. The 22 PBDEs were randomly divided to two groups: Group I, which comprises 16 PBDEs, and Group II, which comprises 6 PBDEs (marked by asterisk in Tables 1 and 2).
MLR model
Generally, a simple model should always be chosen in preference to a complex model, if the latter does not fit the data better. Thus, we firstly investigate whether MLR can model the quantitative relationship between the MDEV index and the lgKOA of these PBDEs. The MDEV index was used as independent variable and the lgKOA was used as dependent variable to develop the model.
Firstly, leave one out cross validation was carried out. In the leave one out cross validation, the lgKOA of all the samples in Group I was predicted in turn. The prediction procedure was performed 16 times. In each time, one sample was selected and used as the test set. The remaining 15 samples were used as training set to develop the regression model. The lgKOA of the selected sample (test set) was then predicted with the obtained regression model. The result of leave one out cross validation is listed in Table 2. As shown in Table 2, the predicted lgKOA are in good agreement with the experimental lgKOA. For the 16 samples of Group I, the prediction RMSRE is 2.82. In addition, the predicted lgKOA were plotted versus the experimental lgKOA. The obtained plot is shown in Figure 1. The plot shows a linear relationship (lgKOA,pred = 0.9635 lgKOA,exp + 0.3573 with R = 0.9769) between the predicted and experimental lgKOA.
Subsequently, external validation was carried out to further assess the predictive ability of the MLR model. The regression model was developed by using all the 16 compounds in Group I. The obtained regression equation is:
The R, Standard error of the estimate and F value of the regression model is 0.9844, 0.2340 and 202.46, respectively. Then, the lgKOA of the six PBDEs in Group II was predicted by Equation 1. The prediction result is shown in Table 2 also. As shown in the table, the predicted lgKOA are still in good agreement with the experimental lgKOA. The prediction RMSRE of the 6 PBDEs in Group II (marked by asterisk in Table 2) is 2.95. The plot of the predicted lgKOA versus experimental lgKOA is presented in Figure 1. As shown in Figure 1, there is a linear relationship (lgKOA,pred = 0.9721 lgKOA,exp + 0.2867 with R = 0.9836) between the predicted and experimental lgKOA.
The results of leave one out cross validation and external validation demonstrates that the MDEV index is quantitatively related to the KOA of PBDEs. The established MLR model can describe the quantitative relationship between the MDEV index and KOA of PBDEs. Compared with the QSPR models reported in the references [12–15], the obtained MLR model shows comparative prediction accuracy. MDEV index can be generated easier than quantum chemical descriptors. Thus, the developed MLR model is a reliable and easy-to-use QSPR model for predicting the KOA of PBDEs.
L-ANN model
L-ANN is an efficient and commonly used multivariate calibration method. Thus, we investigated whether a better model can be developed by using L-ANN appraoch. A 2-1 RBF-ANN (i.e. there are 2 nodes in the input layer and 1 node in the output layer) was used to model the quantitative relationship between the MDEV index and the lgKOA. The MDEV index was used as the input variable and the lgKOA was used as the output variable.
Group I was still used to carry out leave one out cross validation. In the leave one out cross validation, the lgKOA of all the samples in Group I was predicted in turn. The prediction procedure was performed 16 times. In each time, one sample was selected and used as the test set. The remaining 15 samples were used as the calibration set to develop the network. Hence the 15 samples were randomly divided into a training set which includes 12 samples and a verification set which includes 3 samples. The lgKOA of the selected sample (test set) was then predicted with the obtained network. The result of leave one out cross validation is listed in Table 2. For the 16 samples of Group I, the prediction RMSRE is 2.55. The plot of the predicted lgKOA versus the experimental lgKOA is presented in Figure 2. The regression equation and correlation coefficient between the predicted and experimental lgKOA is lgKOA,pred = 0.9731 lgKOA,exp + 0.2640 and 0.9812 respectively.
Subsequently, the external validation was carried out by using all the 22 PBDEs. An L-ANN model was developed from the 16 PBDEs in Group II. In the training procedure, the verification set comprises three randomly selected samples and the rest 13 samples were used as the training set. The lgKOA of the six PBDEs in Group I was then predicted with the obtained L-ANN model. The prediction result is presented in Table 2 also. The prediction RMSRE of the 6 PBDEs in Group II (marked by asterisk in Table 2) is 2.68. The plot of the predicted lgKOA versus the experimental lgKOA is shown in Figure 2. There is a linear relationship (lgKOA, pred = 0.9854 lgKOA, exp + 0.1535 with R =0.9864) between the predicted and experimental lgKOA. Obviously, the predicted lgKOA is in good agreement with the experimental lgKOA. It is demonstrated that the quantitative relationship between the MDEV index and lgKOA of PBDEs has been modeled well by L-ANN. Compared with the QSPR models reported in the references [12–15], the obtained L-ANN model shows comparative accuracy in predicting the lgKOA of PBDEs. Obviously, it is a reliable and easy-to-use QSPR model for predicting the lgKOA of PBDEs. In addition, the prediction result of the L-ANN model is slightly better than the result of the MLR model. Therefore, the established L-ANN model should be a more promising model for studying the octanol/air partition behavior of PBDEs.
Experimental
Data set
The MDEV index was calculated according to the approach presented in section “Methods: MDEV index”. The calculated MDEV index is listed in Table 1. The experimental lgKOA of the 22 PBDEs listed in Table 2 is taken from references [12].
Root mean square relative error (RMSRE) was calculated to indicate the prediction performance of the obtained models. RMSRE is defined as:
where RE i is the relative error of the ith sample, and n is the number of samples.
Software
All the calculations were done with the subroutines developed under Matlab (Ver. 7.0). The computation was performed on a personal computer equipped with an i5-2450M processor. The used activation function of L-ANN is a linear function shown in Equation 5.
Conclusion
Two QSPR models for the octanol/air partition of PBDEs were developed by using MLR and L-ANN respectively. The results of leave one out cross validation and external validation indicate that the obtained MLR model and L-ANN model are practicable for predicting the KOA of PBDEs. It is demonstrated that the MDEV index is quantitatively related to the KOA of PBDEs. MDEV index can be generated easier than quantum chemical descriptors. Thus, using MDEV index as structural descriptor is more convenient than using quantum chemical descriptor when developing the QSPR model for the KOA of PBDEs. In addition, the result demonstrates MLR and L-ANN are both practicable for modeling the quantitative relationship between the MDEV index and KOA of PBDEs. Compared with the established MLR model, the obtained L-ANN model shows slightly higher prediction accuracy. The obtained L-ANN model should be a more promising model for studying the octanol/air partition behavior of PBDEs.
Methods
MDEV index
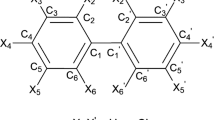
In the calculation of MDEV index, a molecule is regarded as a geometric graph. Each non-hydrogen atom is regarded as a point and each chemical bond is regarded as an edge. The molecular structure of PBDEs can be encoded by the MDEV index of bromine atoms and benzene rings. If the relative electronegative of each bromine atom and benzene ring is defined as 1, the MDEV index of PBDEs can be defined as Equation 3:
(k, l =1,2 and l ≥ k)
where k and l denote the type of an atom (k =1 or l =1 denotes the bromine atom, and k =2 or l =2 denotes the benzene ring); i and j are the coding number of series number of a bromine atom or benzene ring in the molecular skeleton graph. In addition, i and j belong to the kth and lth type respectively. The d ik , jl means the shortest relative distance between the ith and jth atom. For example, di1,j1 denotes the nearest relative distance between the ith and jth bromine atom. The relative bond length between the two adjacent non-hydrogen atoms is defined as d = 1. According to Equation 3, there are three elements, M11, M12 and M22, in the MDEV index for a PBDE molecule. The three elements are usually noted as μ1, μ2 and μ3 respectively. For example, the MDEV index of 2,2’,4,4’-PBDE should be calculated as follows:
Obviously, the M22 of each PBDE is equal to 1. Thus, μ1 and μ2 were used to describe the structure of PBDEs.
Artificial neural network
The theory of ANN has been elaborated in a lot of articles [21–28]. Hence, only a brief outline of ANN is presented here.
ANN is a multivariate calibration method capable of modeling complex functions. The basic processing unit of ANN is the neuron (node). An artificial neural network comprises a number of neurons organized in different layers. Linear artificial neural network (L-ANN) [22–25] is a neural network having no hidden layers, but an output layer with fully linear neurons (that is, linear neurons with linear activation function). It is the simplest artificial neural network. In L-ANN, the neurons between the input and output layers fully connect, while the neurons in the same layer do not. Figure 3 illustrates the basic architecture of the used L-ANN.
In Figure 3, x1 and x2 are the input variables; y1 and w1 denotes the output variables and the element of connection weight matrix W respectively; b1 is the bias vector. The symbol fact( ) means the activation function. Previous to training procedure, the input and output variables are normalized. When the network is executed, it multiplies the input variables by the weights matrix, and then adds the bias vector. The post synaptic potential (PSP) function of the neuron can be described as Equation 5:
Generally, the activation function used in L-ANN is a linear function which can be described as:
Because there are no non-linear functions and hidden neurons in the network, L-ANN is ideal for dealing with linear problems. Actually, training a linear network means finding the optimal setting for the weight matrix W to minimize the root mean squared error (RMSE) of calibration set. In order to achieve this aim, the known samples which are used as calibraion set are generally divided into two parts: a training set and a verification set. The training set was used to calculate and adjust the network weights. The verification set was used to track the network's error performance, to identify the best network, and to stop training. The training should be stopped once deterioration in the verification error is observed. The optimal network parameters were selected according to the RMSE of verification set. The over-fitting and over-learning can be effectively avoided in this way. Although the verification set is used to identify the best network, actually, training algorithms do not use the verification set to adjust network weights. Standard pseudo-inverse linear optimization algorithm [22] is usually used to train the network. This algorithm uses the singular value decomposition technique to calculate the pseudo-inverse of the matrix needed to set the weights in a linear output layer, so as to find the least mean squared solution. Essentially, it guarantees to reach the optimal setting for the weights in the linear layer.
The main difference between MLR and L-ANN is the optimization algorithm. In MLR, the aim of least square algorithm is to minimize the sum of squared residuals of the training set. As for L-ANN, the aim of training algorithm is to minimize the RMSE of verification set [22].
Leave one out cross validation
Leave one out cross validation [29] is a commonly used algorithm for estimating predictive performance of a multivariable calibration model. Usually, practical calibration experiments have to be based on a limited set of available samples. The idea behind the leave one out cross validation algorithm is to predict the property value of each sample in turn with the calibration model which is developed with the other samples. When applying the algorithm to a dataset with N samples, the calibration modeling is performed N times, each time using (N-1) samples for modeling and one sample for testing. Thus, the procedure of leave one out cross validation can be divided into N segment. In each segment i (i = 1, . . . , N), there are three steps: (1) taking sample i out as temporary ‘test set’, which is not used to develop the calibration model, (2) developing the calibration model with the remaining (N-1) samples, (3) testing the developed model with sample i, calculating and storing the prediction error of the sample.
External validation
External validation [26, 30] is a algorithm which has been generally applied to estimating predictive performance of calibration models. When utilizing the algorithm, working dataset is split into two subsets: a calibration set, which is used to establish the calibration model, and a test set, which is employed to assess the predictive ability of the established calibration model. Herein, test set is designed to give an independent assessment of the predictive performance of the assed model. It is not used in establishing the calibration mdoel at all, and hence is independent of the calibration set. Generally, the samples in calibration set and test set are randomly selected from the working dataset.
References
Anderson HA, Imma P, Knobeloch L, Turyk M, Mathew J, Buelow C, Persky V: Polybrominated diphenyl ethers (PBDE) in serum: Findings from a US cohort of consumers of sport-caught fish. Chemosphere. 2008, 73: 187-194. 10.1016/j.chemosphere.2008.05.052.
EPA: America’s children and the environment. 2014, http://www.epa.gov/ace/pdfs/Biomonitoring-PBDEs.pdf,
Secretariat of the Stockholm Convention: Listing of POPs in the Stockholm Convention. 2014, http://chm.pops.int/TheConvention/ThePOPs/ListingofPOPs/tabid/2509/Default.aspx,
Yue CY, Li LY: Filling the gap: estimating physicochemical properties of the full array of polybrominated diphenyl ethers (PBDEs). Environ Pollut. 2013, 180: 312-323.
Wang YW, Zhao CY, Ma WP, Liu HX, Wang T, Jiang GB: Quantitative structure-activity relationship for prediction of the toxicity of polybrominated diphenyl ether (PBDE) congeners. Chemosphere. 2006, 64: 515-524. 10.1016/j.chemosphere.2005.11.061.
Watkins DJ, McClean MD, Fraser AJ, Weinberg J, Stapleton HM, Webster TF: Associations between PBDEs in office air, dust, and surface wipes. Environ Int. 2013, 59: 124-132.
Harner T, Shoeib M: Measurements of octanol-air partition coefficients (KOA) for polybrominated diphenyl ethers (PBDEs): predicting partitioning in the environment. J Chem Eng Data. 2002, 47: 228-232. 10.1021/je010192t.
Mizukawa K, Takada H, Takeuchi I, Ikemoto T, Omori K, Tsuchiya K: Bioconcentration and biomagnification of polybrominated diphenyl ethers (PBDEs) through lower-trophic-level coastal marine food web. Mar Pollut Bull. 2009, 58: 1217-1224. 10.1016/j.marpolbul.2009.03.008.
Wania F, Lei YD, Harner T: Estimating octanol-air partition coefficients of nonpolar semivolatile organic compounds from gas chromatographic retention times. Anal Chem. 2002, 74: 3476-3483. 10.1021/ac0256033.
Han SY, Liang C, Qiao JQ, Lian HZ, Ge X, Chen HY: A novel evaluation method for extrapolated retention factor in determination of n-octanol/water partition coefficient of halogenated organic pollutants by reversed-phase high performance liquid chromatography. Anal Chim Acta. 2012, 713: 130-135.
Cetin B, Odabasi M: Atmospheric concentrations and phase partitioning of polybrominated diphenyl ethers (PBDEs) in Izmir, Turkey. Chemosphere. 2008, 71: 1067-1078. 10.1016/j.chemosphere.2007.10.052.
Xu HY, Zou JW, Yu QS, Wang YH, Zhang JY, Jin HX: QSPR/QSAR models for prediction of the physicochemical properties and biological activity of polybrominated diphenyl ethers. Chemosphere. 2007, 66: 1998-2010. 10.1016/j.chemosphere.2006.07.072.
Wang ZY, Zeng XL, Zhai ZC: Prediction of supercooled liquid vapor pressures and n-octanol/air partition coefficients for polybrominated diphenyl ethers by means of molecular descriptors from DFT method. Sci Total Environ. 2008, 389: 296-305. 10.1016/j.scitotenv.2007.08.023.
Chen JW, Harner T, Yang P, Quan X, Chen S, Schramm KW, Kettrup A: Quantitative predictive models for octanol–air partition coefficients of polybrominated diphenyl ethers at different temperatures. Chemosphere. 2003, 51: 577-584. 10.1016/S0045-6535(03)00006-7.
Papa E, Kovarich S, Gramatica P: Development, validation and inspection of the applicability domain of QSPR models for physicochemical properties of polybrominated diphenyl ethers. QSAR Comb Sci. 2009, 28: 790-796. 10.1002/qsar.200860183.
Nandi S, Monesi A, Drgan V, Merzel F, Novič M: Quantitative structure-activation barrier relationship modeling for Diels-Alder ligations utilizing quantum chemical structural descriptors. Chem Cent J. 2013, 7: 171-10.1186/1752-153X-7-171.
Steffen A, Apostolakis J: On the ease of predicting the thermodynamic properties of beta-cyclodextrin inclusion complexes. Chem Cent J. 2007, 1: 29-10.1186/1752-153X-1-29.
Gutman I, Tosovic J: Testing the quality of molecular structure descriptors. Vertex–degree based topological indices. J Serb Chem Soc. 2013, 78: 805-810. 10.2298/JSC121002134G.
Liu HH, Xiao X, Qin J, Liu YM: Study on structural characteristics and QSPR of polychlorinated biphenyls Isomers (PCBs). J Chongqing Inst Tech (In Chinese). 2005, 19: 67-70.
Liu SS, Liu HL, Xia ZN, Cao CZ, Li ZL: Molecular distance-edge vector (μ): an extension from alkanes to alcohols. J Chem Inf Comput Sci. 1999, 39: 951-957. 10.1021/ci990011f.
Yin CS, Guo WM, Lin T, Liu SS, Fu RQ, Pan ZX, Wang LS: Application of wavelet neural network to the prediction of gas chromatographic retention indices of alkanes. J Chin Chem Soc. 2001, 48: 739-749.
Statsoft: Model extremely complex functions neural networks. 2013, http://www.statsoft.com/textbook/neural-networks,
Yin CS, Shen Y, Liu SS, Yin QS, Guo WM, Pan ZX: Simultaneous quantitative UV spectrophotometric determination of multicomponents of amino acids using linear neural network. Comput Chem. 2001, 25: 239-243. 10.1016/S0097-8485(00)00097-8.
Zhang WJ, Zhong XQ, Liu GH: Recognizing spatial distribution patterns of grassland insects: neural network approaches. Stoch Environ Res Risk Assess. 2008, 22: 207-216. 10.1007/s00477-007-0108-3.
Zhang YX, Li H, Hou AX, Havel J: Artificial neural networks based on principal component analysis input selection for quantification in overlapped capillary electrophoresis peaks. Chemometr Intell Lab Syst. 2006, 82: 165-175. 10.1016/j.chemolab.2005.08.012.
Jalali-Heravi M, Garkani-Nejad Z: Prediction of electrophoretic mobilities of alkyl- and alkenylpyridines in capillary electrophoresis using artificial neural networks. J Chromatogr A. 2002, 971: 207-215. 10.1016/S0021-9673(02)01043-9.
Fatemi MH, Baher E: Quantitative structure-property relationship modelling of the degradability rate constant of alkenes by OH radicals in atmosphere. SAR QSAR Environ Res. 2009, 20: 77-90. 10.1080/10629360902726700.
Abdollahi Y, Zakaria A, Abbasiyannejad M, Masoumi HRF, Moghaddam MG, Matori KA, Jahangirian H, Keshavarzi A: Artificial neural network modeling of p-cresol photodegradation. Chem Cent J. 2013, 7: 96-10.1186/1752-153X-7-96.
Martens HA, Dardenne P: Validation and verification of regression in small data sets. Chemometr Intell Lab Syst. 1998, 44: 99-121. 10.1016/S0169-7439(98)00167-1.
Yin CS, Shen Y, Liu SS, Yin QS, Guo WM, Pan ZX: Simultaneous quantitative UV spectrophotometric determination of multicomponents of amino acids using linear neural network. Compu Chem. 2001, 25: 239-243. 10.1016/S0097-8485(00)00097-8.
Acknowledgments
The work was supported by the National Natural Science Foundation of China No. 21305108, the Project Supported by Natural Science Basic Research Plan in Shaanxi Province of China (Program No. 2014JM2039), and the Innovative Research Team of Xi’an Shiyou University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LJ conceived of the study, carried out the calculation of structural descriptor and the model building, draft the manuscript. MMG and XFW participated in the design of the study and performed the statistical analysis, HL coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits use, duplication, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Jiao, L., Gao, M., Wang, X. et al. QSPR study on the octanol/air partition coefficient of polybrominated diphenyl ethers by using molecular distance-edge vector index. Chemistry Central Journal 8, 36 (2014). https://doi.org/10.1186/1752-153X-8-36
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1752-153X-8-36