Abstract
Background
Staphylococcus aureus is a common bacterium usually found on skin and mucous membranes of warm blooded animals. Resistance in S. aureus has been increasingly reported though depending on the clonal lineage. Indeed, while hospital acquired (HA)-methicillin resistant S. aureus (MRSA) are typically multi-resistant, community associated (CA)-MRSA are by large more susceptible to many antibiotics. Although S. aureus isolated from animals are often susceptible to most antibiotics, multi-resistant livestock associated (LA)-MRSA have been recovered from bovine mastitis.
In this study, we investigated the prevalence and types of MRSA present in the nose of healthy bovines of different age groups and rearing practices. Since no validated methods for MRSA isolation from nasal swabs were available, we compared two isolation methods. Molecular characterization was performed by means of spa-typing, MLST, SCCmec typing and microarray analysis for the detection of antimicrobial resistance and virulence genes.
Results
MRSA between herd prevalence in bovines was estimated at 19.8%. There was a marked difference between rearing practices with 9.9%, 10.2% and 46.1% of the dairy, beef and veal calve farms respectively being MRSA positive. No significant difference was observed between both isolation methods tested. Most isolates were ST398 spa type t011 or closely related spa types. Few ST239 spa type t037 and t388 and ST8 spa type t121 were also found. SCCmec types carried by these strains were mainly type IV(2B), IV(2B&5) and type V. Type III and non-typeable SCCmec were recovered to a lesser extent. All isolates were multi-resistant to at least two antimicrobials in addition to the expected cefoxitin and penicillin resistance, with an average of resistance to 9.5 different antimicrobials. Isolates selected for microarray analysis carried a broad range of antimicrobial resistance and virulence genes.
Conclusion
MRSA were mainly present in veal farms, compared to the lower prevalence in dairy or beef farms. Multi-resistance in these strains was high. Though mainly CC398 spa t011 was found, the genetic diversity was higher than what was found for pigs in Belgium. CC8 strains, a typically human lineage but also recently found also in association with bovines, has been retrieved here also.
Similar content being viewed by others
Background
Staphylococcus aureus is a common facultative pathogenic bacterium that has long been recognized as a burden in both human and veterinary medicine. Indeed, S. aureus has been shown to be responsible of various infections such as clinical and subclinical bovine mastitis [1, 2], wound infections in horses [3–5], dogs [6] and wild animals such as hedgehogs [7]. Furthermore, S. aureus is well known to harbour resistance to antimicrobial agents which may lead to complications in the treatment of its infections [8] and increase the cost of treatments [9]. One of these antimicrobial resistances is encoded by the mecA gene conferring resistance to almost all β-lactams including methicillin, oxacillin and cephalosporins. Though first considered not causing many infections [10], MRSA have more recently been shown to be present in 10% of Belgian farms suffering from S. aureus bovine mastitis [11]. Livestock associated (LA)-MRSA was first described in pigs in 2005 and humans in close contact with pigs in the Nederland [12] and in France [13]. This particular clone belonging to the clonal complex (CC)398 was later encountered in many healthy animals such as pigs [14], horses [15], bovines [16] and poultry [17–19]. This clonal complex is composed of different closely related spa types [20] and cannot be typed by pulsed field gel electrophoresis using SmaI digestion [21].
Although MRSA in bovines and in cases of bovine mastitis are well documented, information about the prevalence of S. aureus and MRSA in healthy bovines is lacking.
For international comparisons, a standardized isolation method is necessary. The European Food Safety Authority (EFSA) [22] has proposed a standardized protocol for the isolation of MRSA from dust samples obtained from pig farms. However, this protocol was estimated not to be very sensitive in a study in poultry in 2011 [19].
The aim of this study was to determine the prevalence and epidemiology of MRSA in bovines and compare the EFSA proposed isolation method with an alternative enrichment method in order to determine whether there were differences between the two methods in this population.
Methods
Sampling and isolation method
Four hundred and thirty-two farms were examined during the national survey on bovine MRSA in Belgium 2012. These farms were randomly selected from the Sanitel database. Of these, 141 were dairy farms, 187 farms reared beef cattle and 104 reared veal calves. Per farm, nose swabs were taken from 20 animals and pooled. Sampling was performed by the Belgian Federal Agency for the Safety of the Food Chain. Ethics approval was not required for this study under Belgian regulations, as taking a nasal swab does not cause pain, distress or lasting harm.
The first method was the standard method proposed by ESFA [22], MRSA was isolated using 100 mL Mueller-Hinton (MH) broth (Becton Dickinson, US) supplemented with NaCl (6.5%) and incubated at 37°C for 20 to 24 h. One ml of this broth was added to Tryptic Soy Broth (TSB) supplemented with cefoxitin (3.5 mg/l) and aztreonam (75 mg/l) and incubated overnight at 37°C. Ten μl of this broth was plated on MRSA selective plate, MRSA-ID (bioMérieux, Marcy-l’Etoile, France), and incubated 48 hours at 37°C. At both 24 and 48 hours, plates were inspected and suspected colonies were purified on Columbia agar plates with 5% sheep blood (CSB) (Bio Rad Laboratories, Nazareth Eke, Belgium) and incubated overnight at 37°C. Since this isolation method includes two enrichment steps, it is referred in this study as double broth enrichment method (DBEM).
The alternative method was applied to 106 farms and differed from the DBEM protocol by the omission of the second enrichment in antibiotic supplemented broth. For this reason, this second isolation methods is referred as single broth enrichment method (SBEM).
DNA extraction, MRSA identification and characterization
DNA was extracted as previously described [3]. MRSA identification and mecA gene detection was performed using a triplex PCR previously published [23].
A PCR allowing the detection of the clonal complex (CC) 398 was performed on all MRSA following a protocol previously described by Stegger et al.[24]. MRSA isolates that were negative in the CC398 PCR were subjected to multi-locus sequence typing (MLST) [25]. Sequences of seven internal fragments were then compared to the international database [26] to obtain the sequence type. Strains were further characterised by spa–typing, as previously described [27]. The resulting spa types were assigned by using the Ridom StaphType software [28]. Clustering of spa types was performed using the algorithm Based Upon Repeat Pattern (BURP) available in the Ridom StaphType software. Staphylococcal cassette chromosome mec (SCCmec) types were determined by the means of two multiplex PCRs (M-PCRs) designed for the detection of the mec-complex and ccr-complex [29]. Appropriate control strains were used.
Antimicrobial susceptibility testing
Antimicrobial resistance was determined using a micro broth dilution method (Sensititre, Trek Diagnostic Systems, Magellan Biosciences, Ohio, USA). The Minimal inhibitory concentrations (MIC) of 19 antimicrobials (penicillin, cefoxitin, kanamycin, streptomycin, gentamicin, erythromycin, clindamycin, quinupristin/dalfopristin, linezolid, tiamulin, chloramphenicol, rifampicin, ciprofloxacin, fusidic acid, tetracycline, trimethoprim, sulfamethoxazole, vancomycin, and mupirocin) were determined as previously described [20]. The MIC values were interpreted using the European Committee on Antimicrobial Susceptibility Testing (EUCAST) epidemiological cut-offs (ECOFF) for S. aureus. Data from the EUCAST MIC distribution website was last accessed November 6, 2013 [30].
DNA microarray-based typing and detection of resistance and virulence genes
Fourteen isolates were selected based on the different antimicrobial resistance phenotypes for detection of resistance and virulence genes by the mean of microarray analysis. Microarray analysis was performed on these strains using the Identibac S. aureus Genotyping DNA Microarray (Alere Technologies GmbH, Köln, Germany) according to the manufacturer’s instructions. The DNA microarray covers 333 oligonucleotide probes, detecting resistance and virulence genes. A full list including primer and probe sequences is available online [31].
Statistical analysis
The number of resistant isolates was counted and resistance percentages were calculated. The Cohen's kappa coefficient was calculated in order to compare both isolation methods. Cohen’s kappa coefficient was interpreted according to Landis & Koch [32]. This analysis includes the first 106 farms. DBEM is considered as the Gold standard while SBEM is the one under estimation. Apparent prevalence, true prevalence, Cohen’s Kappa coefficient, sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of both methods were also calculated using a previously described formulae [33] and Win Episcope 2.2 (Clive, United Kingdom). Pearson chi square and Fisher’s exact test were computed using IMB SPSS Statistics® Version 20.0.
Results
Prevalence
Overall, and using the official DBEM, 81 farms (19.8%, 95% confidence interval (CI) [15.1% - 22.4%]) were positive for MRSA (Table 1). There was no significant difference between the between-herd prevalence of dairy and beef farm (Fisher’s exact test; p = 0.549) while there was a significant difference between the between-herd prevalence of dairy and veal calf farms (Fisher’s exact test; p < 0.001) and between the between-herd prevalence of beef and veal calf farms (Fisher’s exact test; p < 0.001). Fourteen dairy farms (9.9%, 95% CI [5.0% - 14.9%]), 19 farms holding beefs (10.2%, 95% CI [5.8% - 14.5%]) and 48 farms rearing veal calves (46.1%, 95% CI [36.6% - 55.7%]) were found positive for MRSA.
Comparison of isolation methods
Comparisons were performed on 106 samples. Using both isolation methods (Table 2), 34 (32.1%, 95% CI [23.2% - 41.1%]) farms out of 106 tested were found to be positive. Among these positive farms recovered, nine farms were detected positive with the SBEM but not with the DBEM and conversely, nine other farms were detected positive with the DBEM but not with the SBEM. Kappa agreement coefficient (k) was 0.61 which indicates a substantial agreement between both methods. There was no significant difference between the prevalence of these methods (Fisher’s exact test; p =0.597). Specificity, positive predictive value and negative predictive value were likewise identical (Table 3).
MLST, spa- and SCCmectyping
Among 81 MRSA isolates recovered, seventy-eight (96.3%) were positive in the CC398 PCR. The three other isolates were ST8 and two ST239, as demonstrated by MLST. All calf isolates were CC398. The ST8 was recovered from beef cattle and both ST239 isolates were isolated from dairy farms (Table 1).
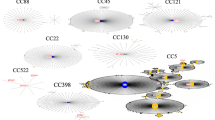
Ten different spa types were identified. Sixty-four (79.0%) were spa type t011. Other spa types recovered were t037 (n = 1), t121 (n = 1), t388 (n = 1), t1451 (n = 3), t1456 (n = 3), t1985 (n = 4), t3423 (n = 1), t6228 (n = 2) and a non-typeable spa type. Two clusters were distinguished using the BURP algorithm (Figure 1). The first cluster, including 92% of all isolates and 44% of all spa types, grouped the spa types t011, t1451, t1456 and t1985. The second cluster, which included 3% of all isolates and 22% of all spa types, grouped the spa types t037 and t388. A singleton was also detected with the spa-type t121. The remaining spa types t3423 and t6228 could not be aligned by the software. All t011 and closely related spa-type isolates were associated to CC398. MRSA spa type t121 was associated to MLST type ST8, while t388 and t037 to ST239. The MRSA t011 and closely related strains were isolated from veal (n = 47), beef (n = 18) and dairy farms (n = 9). The t3423 and t6228 MRSA were isolated from veal (n = 1) and dairy farms (n = 2). The t037, t388 and the non-typable spa type MRSA were recovered from dairy farms and the t121 MRSA was recovered in beef farm (Table 1).
Forty-four (54.3%) isolates carried SCCmec type IV(2B) and nine (11.1%) SCCmec type IV(2B&5). Sixteen (19.8%) isolates carried SCCmec type V(5C2) and two (2.5%) SCCmec type III(3A). Ten (12.3%) isolates showed only the mecA gene but no ccr complex was detected with the PCR. These were thus considered non-typeable using these M-PCRs. SCCmec type IV (2B and/or 2B&5) were found in isolates from veal (n = 37), beef (n = 12) and dairy farms (n = 4). SCCmec type V were also found in the three age groups with seven being found in isolates from veal, six from dairy and three from beef cattle. Type III cassette were found in from dairy (n = 1) and beef cattle (n = 1). The non-typeable SCCmec was detected in isolates from veal calves (n = 4), dairy (n = 3) and beef (n = 3) cattle. Additionally to the type IV(2B) (n = 43), IV(2B&5) (n = 9), V (n = 16) and non-typeable (n = 8) SCCmec, CC398 MRSA isolated also carried the type III (n = 2) SCCmec. Both t121 and the non-typeable spa type carried SCCmec type IV(2b) and spa types t388 and t037 carried a non typeable SCCmec.
Antimicrobial resistance
All isolates were resistant to cefoxitin and penicillin as expected. More than 90% of the isolates were resistant to tetracycline (96.3%) and trimethoprim (95.1%). A high prevalence of resistance was also observed to clindamycin (86.4%), erythromycin (86.4%), kanamycin (80.2%) and gentamicin (76.5%). More than half of the isolates were also resistant to streptomycin (58.0%). Lower resistance levels were detected to fusidic acid (27.2%), sulfamethoxazole (25.9%), quinupristin/dalfopristin (23.5%), tiamulin (17.3%), ciprofloxacin (16.0%), chloramphenicol (12.3%), rifampicin (12.3%) and mupirocin (9.9%). No resistance was observed to linezolid and vancomycin (Table 4). All isolates were at least resistant to two more antimicrobials in addition to cefoxitin and penicillin. More than 50% of the isolates were resistant to nine or more different antimicrobials. Two isolates were resistant to 16 different antimicrobials, remaining susceptible only to ciprofloxacin, linezolid and vancomycin. The isolates resistant to 15 (n = 3) or 16 (n = 2) antibiotics were all CC398 spa type t011. Two of these isolates carried a non-typable cassette and three carried SCCmec type IV (2B). These originated from veal (n = 3) and beef cattle (n = 2). The one isolate resistant to 14 antibiotics was a CC398 spa type t6228 strain carrying SCCmec type V. The one isolate resistant to only four antibiotics was a CC398 spa type t1456 strain carrying SCCmec type V and originated from a farm holding beef cattle. Isolates that were resistant to five (n = 1) and six (n = 5) antimicrobials were CC398 spa type t011 carrying SCCmec type V (n = 3) and IV (2B; n = 1) or t1985 (n = 2). These isolates were isolated from veal calves (n = 3), dairy (n = 1) and beef cattle (n = 2). The ST8 isolate was resistant to seven different antimicrobials and both ST239 isolates were resistant to nine different antimicrobials. There were no significant differences in resistance prevalence between veal calves, dairy and beef cattle.
Microarray typing for resistance and virulence gene detection
Most genes were homogeneously distributed in all isolates, including typical S. aureus species marker and regulatory genes (23S-rRNA, gapA, katA, coA, nuc, spa, sbi, sarA, saeS, vraS), the accessory gene regulator agrI, haemolysins (hla, hld), genes encoding leukocidins (lukS-F, hlgA, lukX, lukY-variant 1), proteases (aur, sspA, sspB, sspP), the biofilm production genes of the icaACD operon, adhesion factors (bbp, cflA, cflB, ebh, ebpS, eno, fib, fnbA, fnbB, map, sdrC, sdrD, vwb) immune-evasion factors (isaB, isdA, hysA1, hysA2), a putative transport protein (lmrP), a site specific deoxyribonuclease subunit X (hsdSx), and staphylococcal superantigen-like proteins from the vSaα genomic islands [setB1, setB2, setB3, setC, ssl1 (set6), ssl2 (set7), ssl4 (set9/ssl4), ssl5 (set3/ssl5), ssl7 (set1/ssl7) and ssl10 (set4/ssl10)].
All isolates were penicillin resistant and carried the bla operon (blaZ, blaI, and blaR) encoding for penicillin-ampicillin resistance. All isolates, except the tetracycline sensitive one, carried the tetracycline resistance gene tetM. Additionnaly to tetM, isolates harbouring SCCmec type V and a non-typeable isolate carried also the tetracycline resistance gene tetK. Six erythromycin resistant isolates out of 11 carried the ermC gene. Eight gentamicin resistance isolates out of the nine tested showed the aminoglycoside adenyl‒/phosphotransferase encoding gene aacA-aphD. Eight kanamycin resistant isolates out of 11 carried the aadD aminoglycoside resistance gene and one additionally carried aminoglycoside phosphotransferase aphA3. One of the two chloramphenicol resistant isolate carried the cat (pMC524) gene encoding for chloramphenicol acetyltransferase. All isolates carried the putative transport protein sdrM. The metallothiol transferase (fosB) gene encoding fosfomycin resistance was detected in both non CC398 isolates. Furthermore, most isolates carried an intact beta-haemolysin gene (hlb), except the ST239 isolate which harboured the hlb gene truncated after the probable insertion of the immune-evasion phage-borne genes sak (staphylokinase) and scn (staphylococcal complement inhibitor).
Discussion
In this study, we found an estimated MRSA prevalence of 19.8% in bovine farms in Belgium. As found in The Netherlands [16] and a small former Belgian study [34], the prevalence in veal calve farms sampled using nasal swabs was much higher than in dairy farms or farms holding beef cattle. In contrast, the prevalence found at veal calve farms was lower than in these previous studies. In the Netherlands, MRSA prevalence in veal calve farms was estimated at 88% [16] while the small scale Belgian study estimated a prevalence of 64% [34]. In this study, swabs were pooled according to EFSA recommendations [35], while in the other two studies, ten to 25 individual samples per farm were analyzed. The lower prevalence in our study may thus be explained by the differences in sampling since it has been shown that a culture of pooled swabs may have a lower sensitivity than separated swabs culture [36]. Compared to other livestock animals, the estimated prevalence in bovines is much higher than that in poultry (0.8%) [20] but lower than that in pigs (68%) [14].
The isolation method used throughout the study (the DBEM) was the method recommended by EFSA and the European reference laboratory. During the comparison, nine isolates were detected while using the DBEM and not with the alternative method. Conversely nine isolates were detected using the SBEM while not with the DBEM. These isolates have probably been lost during the second enrichment step since this includes antimicrobials. However, as shown for samples from poultry [19], representing a low prevalence population, the second enrichment method does not make any statistical difference. Therefore we recommend for future European surveillances to use the SBEM on nasal swabs.
Most isolates were typical LA-MRSA CC398 spa type t011 or closely related spa types. Three other MLST types were recovered: ST8 spa type t121 and ST239 spa type t037 and t388. Those three types are usually identified among hospital-acquired (HA)-MRSA. However, while MRSA spa type t121 was uncommonly found in Belgian hospitals [37] it has been commonly recovered in hospitals in Europe and in the United States [38]. This spa type has also been found in bulk tank milk in the United States [39]. MRSA ST239 spa type t388 and t037 are widespread HA-MRSA found in Europe, Asia and America [40]. A MRSA ST239 t037 was also isolated from poultry in 2011 [19]. The recovery of these HA-MRSA from livestock indicates that one should remain vigilant to the evolution of MRSA in animals. Though not investigated in his study, these strains in general carry a multitude of virulence genes on mobile genetic elements. Transfer of these virulence genes to LA-MRSA CC398 would have a huge impact on the importance of this clone for human health and its epidemiology in animals.
The diversity of spa types seen in this study in bovines was larger than what has been found previously in pigs in Belgium, where only spa type t011 and t034 were found [14, 41]. In bovines, at least seven different spa types were recovered among the MRSA CC398 isolates. It has been concluded previously that the length of the spa gene sequence may depend on the fact that isolates are methicillin resistance or not, or on the source of S. aureus isolation [42]. Since our isolates were all methicillin resistant and spa-types were found to be closely related, the hypothesis of a possible host adaptation is supported. Also the diversity of the SCCmec types in isolates from cattle seems to be larger than what is found in pigs, however the two predominant types are the same, SCCmec type IV and SCCmec type V. Surprisingly, two isolates carried SCCmec type III. This type is typically associated with HA-MRSA [43] and has also been found extensively in Staphylococcus spp. other than S. aureus from animals. SCCmec type III has been described before in ST398, but these were in fact variant SCCmec type V [44, 45]. Next to this, six isolates carried a non-typeable SCCmec cassette. Further studies are needed to be able to estimate the plasticity of the SCCmec, since this may be of importance to the epidemiology of MRSA in livestock and humans.
The level of multi-resistance is extremely high since it accounts for an average of 9.5 different antimicrobials. Most isolates were resistant to tetracycline and trimethoprim additionally to the expected resistance to cefoxitin and penicillin. In this study two CC398 isolates were found to be susceptible to tetracycline while tetracycline susceptible strains are only very rarely found in CC398 MRSA [46]. The prevalence of erythromycin, clindamycin, kanamycin and gentamicin resistance in this collection is extremely high compared to what has been found in strains from other origins in Belgium [14, 34]. The isolate with the lowest level of multi-resistance was resistant to two additional antimicrobials. Two isolates were resistant to sixteen antimicrobials out of nineteen tested excluding ciprofloxacin, linezolid and vancomycin, three antimicrobials that are used as a last resort in the treatment of MRSA infections in humans.
The staphylokinase (sak) and the staphylococcal complement inhibitor (scn) genes carried on bacteriophage of the φ3 family were found only on the ST239 isolate but not on the most typical LA-MRSA. Since this bacteriophage family are commonly found in human isolates but few in isolates from animals or humans in contact with pigs [47, 48], this might indicate a human to animal transmission of non CC398 isolates. Most resistance and virulence gene detected were homogeneously distributed amongst isolates except for the macrolide/lincosamide resistance encoding gene erm(C) which was found in more than half of the erythromycin resistant isolates and the fosfomycin resistance gene fosB which was detected in two non CC398 isolates. Additionally to resistance genes, virulence factors such as leukocidins, proteases, staphylococcal superantigen like proteins, haemolysins genes, genes involved in adhesion and immune-evasion were also found in all isolates tested by micro-array. Our results are similar to those of a previous micro-array based study performed in Germany [49] on S. aureus isolates from cattle in which leukicidins, haemolysins and enterotoxin genes were detected in most isolates. According to this study, staphylokinase (sak) was also absent in most of our isolates except for the ST239 isolate. However, while in the German study toxic shock syndrome toxins, were demonstrated, the tst-1 gene was not detected in our isolates. Additionally, genes encoding adhesion factors including the bone sialoprotein-binding protein (bbp), the cell wall associated fibronectin‒binding protein (ebh), the fibrinogen binding protein (fib), the fibronectin‒binding protein (fnbB) and the major histocompatibility complex class II analog protein (map) were detected in all isolates. These genes were also found in MRSA isolates from Sahiwal cattle with mastitis in India [50]. Our results show that, although our isolates came from apparently healthy carrier animals, MRSA in bovines may carry a broad range of different resistance genes and virulence factor that might play an important role in the pathogenicity of the bacteria.
Conclusion
In conclusion, MRSA were found in bovines in different rearing practices. Estimated prevalence was, however, lower in nasal isolates from dairy and beef cows than from veal calves. No significant difference was observed between both isolation methods tested. The diversity of strains was larger than what was seen in pigs. Indeed, more different spa-types were recovered in bovine’s isolates than in pigs. Additionally, the diversity in SCCmec cassettes in CC398 was shown not to be limited to the types IV and V but included also type III and a non-typeable cassette. A high level of multi-resistance was found and a broad range of antimicrobial resistance and virulence genes was detected though animals sampled were apparently healthy.
Abbreviations
- CC:
-
Clonal complex
- CHL:
-
Chloramphenicol
- CI:
-
Confidence interval
- CIP:
-
Ciprofloxacin
- CLI:
-
Clindamycin
- CSB:
-
Columbia sheep blood
- DBEM:
-
Double broth enrichment method
- ECOFF:
-
Epidemiological cut-offs
- EFSA:
-
European Food Safety Authority
- ERY:
-
Erythromycin
- EUCAST:
-
European committee on antimicrobial susceptibility Testing
- FASFC:
-
Federal Agency for the Safety of the Food Chain
- FOX:
-
Cefoxitin
- FUS:
-
Fusidic acid
- GEN:
-
Gentamicin
- HA:
-
Hospital-acquired
- k:
-
Kappa agreement coefficient
- KAN:
-
Kanamycin
- LA:
-
Livestock-associated
- LZD:
-
Linezolid
- MH:
-
Mueller-Hinton
- MIC:
-
Minimal inhibitory concentration
- MLST:
-
Multi-locus sequence typing
- MUP:
-
Mupirocin
- M-PCRs:
-
Multiplex PCRs
- MRSA:
-
Methicillin resistant Staphylococcus aureus
- NT:
-
Non-typable
- PEN:
-
Penicillin
- R:
-
Resistance
- RIF:
-
Rifampicin
- SBEM:
-
Single broth enrichment method, SCCmec, Staphylococcal cassette chromosome mec
- SMX:
-
Sulfamethoxazole
- ST:
-
Sequence type
- STR:
-
Streptomycin
- SYN:
-
Quinupristin/dalfopristin
- TET:
-
Tetracyclin
- TIA:
-
Tiamulin
- TMP:
-
Trimethoprim
- TSB:
-
Tryptic Soy Broth
- VAN:
-
Vancomycin.
References
Tenhagen BA, Köster G, Wallmann J, Heuwieser W: Prevalence of mastitis pathogens and their resistance against antimicrobial agents in dairy cows in Brandenburg, Germany. J Dairy Sci. 2006, 89: 2542-2551.
Vanderhaeghen W, Cerpentier T, Adriaensen C, Vicca J, Hermans K, Butaye P: Methicillin-resistant Staphylococcus aureus (MRSA) ST398 associated with clinical and subclinical mastitis in Belgian cows. Vet Microbiol. 2010, 144: 166-171.
Hartmann FA, Trostle SS, Klohnen AA: Isolation of methicillin-resistant Staphylococcus aureus from a postoperative wound infection in a horse. J Am Vet Assoc. 1997, 211: 590-592.
Seguin JC, Walker RD, Caron JP, Kloos WE, George CG, Hollis RJ, Jones RN, Pfaller MA: Methicillin-resistant Staphylococcus aureus outbreak in a veterinary teaching hospital: potential human-to-animal transmission. J Clin Microbiol. 1999, 37: 1459-1463.
van Duijkeren E, Moleman M, van Oldruitenborgh-Oosterbaan MM S, Multem J, Troelstra A, Fluit AC, van Wamel WJ, Houwers DJ, de Neeling AJ, Wagenaar JA: Methicillin-resistant Staphylococcus aureus in horses and horse personnel: an investigation of several outbreaks. Vet Microbiol. 2010, 141: 96-102.
Gortel K, Campbell KL, Kakoma I, Whittem T, Schaeffer DJ, Weisiger RM: Methicillin resistance among staphylococci isolated from dogs. Am J Vet Res. 1999, 60: 1526-1530.
Monecke S, Gavier-Widen D, Mattsson R, Rangstrup-Christensen L, Lazaris A, Coleman DC, Shore AC, Ehricht R: Detection of mecC-Positive Staphylococcus aureus (CC130-MRSA-XI) in Diseased European Hedgehogs (Erinaceus europaeus) in Sweden. PLoS One. 2013, 8: e66166.
Lowy FD: Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest. 2003, 111: 1265-1273.
Huijps K, Lam TJ, Hogeveen H: Costs of mastitis: facts and perception. J Dairy Res. 2008, 75: 113-120.
Devriese LA, Van Damme LR, Fameree L: Methicillin-(cloxacillin)-resistant Staphylococcus aureus strains isolated from bovine mastitis cases. Zentralbl Veterinarmed B. 1972, 19: 598-605.
Voss A, Loeffen F, Bakker J, Klaassen C, Wulf M: Methicillin-resistant Staphylococcus aureus in pig farming. Emerg Infect Dis. 2005, 11: 1965-1966.
Armand- Lefevre L, Ruimy R, Andremont A: Clonal comparison of Staphylococcus aureus isolates from healthy pig farms, human controls and pigs. Emerg Infect Dis. 2005, 11: 711-714.
Baba K, Ishihara K, Ozawa M, Tamura Y, Asai T: Isolation of meticillin-resistant Staphylococcus aureus (MRSA) from swine in Japan. Int J Antimicrob Agents. 2010, 36: 352-354.
Crombé F, Willems G, Dispas M, Hallin M, Denis O, Suetens C, Gordts B, Struelens M, Butaye P: Prevalence and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus among pigs in Belgium. Microb Drug Resist. 2012, 18: 125-131.
Van den Eede A, Martens A, Lipinska U, Struelens M, Deplano A, Denis O, Haesebrouck F, Gasthuys F, Hermans K: High occurrence of methicillin-resistant Staphylococcus aureus ST398 in equine nasal samples. Vet Microbiol. 2009, 133: 138-144.
Graveland H, Wagenaar JA, Heesterbeek H, Mevius D, van Duijkeren E, Heederik D: Methicillin resistant Staphylococcus aureus ST398 in veal calf farming: Human MRSA carriage related with animal antimicrobial usage and farm hygiene. PLoS One. 2010, 5: e10990.
Persoons D, Van Hoorebeke S, Hermans K, Butaye P, de Kruif A, Haesebrouck F, Dewulf J: Methicillin-resistant Staphylococcus aureus in poultry. Emerg Infect Dis. 2009, 15: 452-453.
Geenen PL, Graat EA, Haenen A, Hengeveld PD, Van Hoek AH, Huijsdens XW, Kappert CC, Lammers GA, Van Duijkeren E, Van De Giessen AW: Prevalence of livestock associated MRSA on Dutch broiler farms and in people living and/or working on these farms. Epidemiol Infect. 2013, 141: 1099-1108.
Nemeghaire S, Roelandt S, Argudín MA, Haesebrouck F, Butaye P: Characterization of methicillin-resistant Staphylococcus aureus from healthy carrier chickens. Avian Pathol. 2013, 42: 342-346.
Denis O, Suetens C, Hallin M, Catry B, Ramboer I, Dispas M, Willems G, Gordts B, Butaye P, Struelens MJ: Methicillin-resistant Staphylococcus aureus ST398 in swine farm personnel, Belgium. Emerg Infect Dis. 2009, 15: 1098-1101.
Bens CC, Voss A, Klaassen CH: Presence of a Novel DNA Methylation enzyme in methicillin-resistant Staphylococcus aureus isolates associated with pig farming leads to uninterpretable results in standard pulsed-field gel electrophoresis analysis. J Clin Microbiol. 2006, 44: 1875-1876.
Anon: Scientific Opinion of the Panel on Biological Hazards on a request from the European Commission on Assessment of the Public Health significance of methicillin resistant Staphylococcus aureus (MRSA) in animals and foods. The EFSA J. 2009, 993: 1-73.
Maes N, Magdalena J, Rottiers S, De Gheldre Y, Struelens MJ: Evaluation of a triplex PCR assay to discriminate Staphylococcus aureus from coagulase-negative staphylococci and determine methicillin resistance from blood cultures. J ClinMicrobiol. 2002, 40: 1514-1517.
Stegger M, Andersen PS, Kearns A, Pichon B, Holmes MA, Edwards G, Laurent F, Teale C, Skov R, Larsen AR: Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecALGA251. Clin Microbiol Infect. 2011, 49: 732-734.
Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG: Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000, 38: 1008-1015.
Multi Locus Sequence Typing. http://saureus.mlst.net.
Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, Vogel U: Typing of methicillin-resistant Staphylococcus aureus in a University Hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003, 41: 5442-5448.
Ridom StaphType. http://www.ridom.de/staphtype.
Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Hiramatsu K: Combination of multiplex PCRs for Staphylococcal cassette chromosome mec type assignement: Rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007, 51: 264-274.
European Committee on Antimicrobial Susceptibility Testing. http://www.eucast.org.
Alere Technologies GmbH. http://alere-technologies.com]
Landis JR, Koch GG: The measurement of observer agreement for categorical data. Biometrics. 1977, 33: 159-174.
Dohoo I, Martin W, Stryhn H: Veterinary Epidemiologic Research. 2nd edition. AVC Inc: Charlottetown; 2009.
Vandendriessche S, Vanderhaeghen W, Soares FV, Hallin M, Catry B, Hermans K, Butaye P, Haesebrouck F, Struelens MJ, Denis O: Prevalence, risk factors and genetic diversity of methicillin-resistant Staphylococcus aureus carried by humans and animals across livestock production sectors. J Antimicrob Chemother. 2013, 68: 1506-1510.
Anon: Technical specifications on the harmonised monitoring and reporting of antimicrobial resistance in methicillin-resistant Staphylococcus aureus in food-producing animals and food. EFSA J. 2012, 10: 2897.
Grmek-Kosnik I, Ihan A, Dermota U, Rems M, Kosnik M, Jorn Kolmos H: Evaluation of separate vs pooled swab cultures, different media, broth enrichment and anatomical sites of screening for the detection of methicillin-resistant Staphylococcus aureus from clinical specimens. J Hosp Infect. 2005, 61: 155-161.
Wildemauwe C, De Brouwer D, Godard C, Buyssens P, Dewit J, Joseph R, Vanhoof R: The use of spa and phage typing for characterization of a MRSA population in a Belgian hospital: Comparison between 2002 and 2007. Pathol Biol. 2010, 58: 70-72.
Ridom SpaServer. [http://spa.ridom.de]
Haran KP, Godden SM, Boxrud D, Jawahir S, Bender JB, Sreevatsan S: Prevalence and characterization of Staphylococcus aureus, including methicillin-resistant Staphylococcus aureus, isolated from bulk tank milk from Minnesota dairy farms. J Clin Microbiol. 2012, 50: 688-695.
Campanile F, Bongiorno D, Borbone S, Stefani S: Methicillin-resistant Staphylococcus aureus Evolution – The multiple facets of an old pathogen. Eur Infec Dis. 2010, 4: 70-76.
European Food Safety Authority. http://www.efsa.europa.eu/en/efsajournal/pub/1597.htm]
Shakeri F, Shojai A, Golalipour M, Rahimi Alang S, Vaez H, Ghaemi EA: spa diversity among MRSA and MSSA Strains of Staphylococcus aureus in North of Iran. Int J Microbiol. 2010, doi:10.1155/2010/351397
Moroney S, Heller L, Arbuckle J, Talavera M, Widen R: Staphylococcal cassette chromosome mec and Panton-Valentine leukocidin characterization of methicillin-resistant Staphylococcus aureus clones. J Clin Microbiol. 2007, 45: 1019-1021.
van Loo I, Huijsdens X, Tiemersma E, de Neeling A, van de Sande-Bruinsma N, Beaujean D, Voss A, Kluytmans J: Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg Infect Dis. 2007, 13: 1834-1839.
Jansen MD, Box AT, Fluit AC: SCCmec typing in methicillin-resistant Staphylococcus aureus strains of animal origin. Emerg Infect Dis. 2009, 15: 137.
Kadlec K, Schwarz S: Identification of a novel trimethoprim resistance gene, dfrK, in a methicillin-resistant Staphylococcus aureus ST398 strain and its physical linkage to the tetracycline resistance gene tet(L). Antimicrob Agents Chemother. 2009, 53: 776-778.
Sung JM, Lindsay JA: Staphylococcus aureusstrains that are hypersusceptible to resistance gene transfer from enterococci. Antimicrob Agents Chemother. 2007, 51: 2189-2191.
McCarthy AJ, van Wamel W, Vandendriessche S, Larsen J, Denis O, Garcia-Graells C, Uhlemann AC, Lowy FD, Skov R, Lindsay JA: Staphylococcus aureus CC398 clade associated with human-to-human transmission. Appl Environ Microbiol. 2012, 78: 8845-8848.
Monecke S, Kuhnert P, Hotzel H, Slickers P, Ehricht R: Microarray based study on virulence-associated genes and resistance determinants of Staphylococcus aureus isolates from cattle. Vet Microbiol. 2007, 125: 128-140.
Kumar R, Yadav BR, Singh RS: Antibiotic resistance and pathogenicity factors in Staphylococcus aureus isolated from mastitic Sahiwal cattle. J Biosci. 2011, 36: 175-188.
Acknowledgments
This research was funded by the EMIDA ERA-Net Project “Methicillin-resistant Staphylococcus aureus lineages in primary productions: multi-host pathogen, spill-over and spill-back between animals and humans?”, project acronym LA-MRSA and CODA-CERVA. Dr. M. A. Argudín is supported by a postdoctoral grant from the Fundación Alfonso Martín Escudero.
We are also very grateful to Andy Lucchina, Déborah Petrone and Léna Demazy for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interest
The authors declare that they have no competing interests.
Authors’ contributions
SN performed isolation and identification of isolates, molecular typing and susceptibility testing, analyzed the data and drafted the manuscript. MAA, PB and FH participated in the design of the study provided scientific advice and helped with editing and revision of the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Nemeghaire, S., Argudín, M.A., Haesebrouck, F. et al. Epidemiology and molecular characterization of methicillin-resistant Staphylococcus aureusnasal carriage isolates from bovines. BMC Vet Res 10, 153 (2014). https://doi.org/10.1186/1746-6148-10-153
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1746-6148-10-153