Abstract
Introduction
Neuropathic pain is believed to be influenced in part by inflammatory processes. In this study we examined the effect of variability in the C-type lectin gene cluster (Aplec) on the development of neuropathic pain-like behavior after ligation of the L5 spinal nerve in the inbred DA and the congenic Aplec strains, which carries seven C-type lectin genes originating from the PVG strain.
Results
While both strains displayed neuropathic pain behavior early after injury, the Aplec strain remained sensitive throughout the whole study period. Analyses of several mRNA transcripts revealed that the expression of Interleukin-1β, Substance P and Cathepsin S were more up-regulated in the dorsal part of the spinal cord of Aplec rats compared to DA, indicating a stronger inflammatory response. This notion was supported by flow cytometric analysis revealing increased infiltration of activated macrophages into the spinal cord. In addition, macrophages from the Aplec strain stimulated in vitro displayed higher expression of inflammatory cytokines compared to DA cells. Finally, we bred a recombinant congenic strain (R11R6) comprising only four of the seven Aplec genes, which displayed similar clinical and immune phenotypes as the Aplec strain.
Conclusion
We here for the first time demonstrate that C-type lectins, a family of innate immune receptors with largely unknown functions in the nervous system, are involved in regulation of inflammation and development of neuropathic pain behavior after nerve injury. Further experimental and clinical studies are needed to dissect the underlying mechanisms more in detail as well as any possible relevance for human conditions.
Similar content being viewed by others
Introduction
Injuries to either the peripheral or central nervous systems (CNS) often lead to chronic neuropathic pain conditions. The underlying mechanisms are not clarified in detail, hence therapeutic options are limited. However immune related reactions in the nervous system are suggested to be of importance both for the maintenance and development of neuropathic pain [1, 2]. One such feature is the recruitment of leukocytes into the CNS after a peripheral nerve injury, which may amplify or modify the inflammatory activation of CNS resident glial cells, in turn leading to exaggerated pain [3–6]. Thus, infiltration of blood monocyte-derived macrophages, is an early phenomenon upon nerve injury. Although involved in the clearance of debris due to their phagocytic properties, activated macrophages also release a range of cytokines and chemokines, which have been linked to pain-related behavior [7–9]. In previous studies the chemokine ligand 2 (Ccl2)- chemokine receptor 2 (Ccr2) signaling has been shown to be critically important for the attraction of monocytes to the CNS, which is in turn of relevance for development of neuropathic pain [6]. Also, other types of leukocytes, including T-cells have been suggested to be involved in neuropathic pain-like behavior [5, 10].
Previous studies on inbred rat and mice strains suggest a considerable genetic contribution to various experimental pain phenotypes [11–14]. However, knowledge of exactly defined molecular pathways involved in, or if genetic influence acts on regulation of inflammatory processes of relevance for neuropathic pain, is limited. In an earlier study we could demonstrate that the MHC locus, a region of about 200 genes, exerts a significant effect on pain susceptibility in inbred rat strains after a peripheral nerve injury [15]. Interestingly, we recently replicated this finding in humans by showing that carriers of the HLA DQB1*03:02 allele displayed an increased risk of developing a neuropathic pain condition after a peripheral nerve lesion [16]. The mechanisms underlying this genetic effect are still unclear, but effects on nerve injury-induced immune reactions are likely given the role of the MHC in these contexts.
To further examine the role of genetically regulated immune reactions for pain susceptibility after nerve injury we here investigated the effect of a small rat chromosome 4 gene fragment containing seven C-type lectin receptors (CLRs). The gene cluster, denoted antigen-presenting lectin-like receptor gene complex (Aplec), has previously been studied primarily in models of autoimmune and infectious disease, where it has been demonstrated to regulate different aspects of the innate immune response [17, 18]. Also, we recently found the Aplec cluster to regulate the immune phenotype after a mechanical ventral root injury, including effects on leukocyte recruitment [19]. The aim here was to explore the importance of variability in the Aplec cluster occurring among inbred rat strains for neuropathic pain-like behavior and immune phenotype after a standardized spinal nerve injury.
Results
The Aplec strain is susceptible to develop neuropathic-pain like behavior.
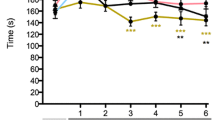
DA and Aplec rats are genetically identical except that the latter contains a small genetic fragment from the PVG strain comprising seven genes, all of which are CLRs (Figure 1) [18, 20, 21]. We here tested susceptibility to develop neuropathic pain behavior in a SNL model. Initially, both strains developed mechanical hypersensitivity to a similar degree after SNL, however, DA rats had started to recover after 14 days, whereas the Aplec rats remained sensitive during the entire time of testing (Figure 2). Statistical analysis demonstrated overall differences between the strains, as well as statistically significant differences specifically on day 21, 28 and 35, the last three time points that were tested.
Neuropathic pain-like behavior after peripheral nerve injury in DA and Aplec strain. One-way ANOVA demonstrates overall differences between the strains (***p < 0.001). Bonferroni post-hoc testing reveals significant difference between DA and Aplec on days 21, 28 and 35 (+++p < 0.001), Data are expressed as means ± SEM. (12–20 rats per group).
Expression of Aplec genes
In order to study any possible expression differences of the seven CLRs in the Aplec fragment the three anatomical locations primarily affected by the injury, i.e. SC, DRG and nerve, were analyzed by RT-PCR. In general, the injury induced expression in the studied tissues was most pronounced in the peripheral nerve, where Dcir1-4 all were increased on both strains (Figure 3A-D). Also in the L5 DRG and the SC these four transcripts were up-regulated after injury but to a lower degree. In contrast, Mcl and Mincle were not significantly affected by injury in any of the locations (Figure 3E-F). In the comparison between strains, Dcir1 and 4 were higher in Aplec compared to DA in peripheral nerve, while Dcir2 was higher in L5 DRG and SC of DA rats. The expression of Dcar1 was below detection limit (data not shown).
Expression levels of Dcir1-4, Mcl and Mincle detected in spinal cord, L4 DRG, L5 DRG and nerve. mRNA levels were analyzed by RT-PCR in Aplec and DA strain 7 days after injury (+) and in healthy controls (−) from each strain. Dcir1 (A), Dcir2 (B), Dcir3 (C), Dcir4 (D), Mcl (E), Mincle (F). One-way ANOVA was done followed by Step-down Bonferroni correction for multiple comparisons (*p < 0.05; **p < 0.01).
Expression of neuropeptides, cytokines and chemokines receptors
The cascade of events occurring after a peripheral nerve lesion includes changes in the expression of neuropeptides, cytokines and chemokines. We therefore measured the expression of Substance P (SP) and Calcitonin gene-related peptide (CGRP), Interleukin-1β (IL1β), Ccr2, Fractalkine receptor (Cx3cr1) and Cathepsin S (CatS). As expected, there was up-regulation of IL-1β expression in all studied tissues (Figure 4A). SP and CGRP were down-regulated in the lesioned L5 DRG, with a trend for higher expression in the adjacent intact L4 DRG (Figure 4B-C). Ccr2, Ccl2, Cx3cr1 and CatS were only studied in the SC, where the latter two were up-regulated (Figure 4D). The expression of IL1β and CatS were higher in SC of Aplec compared to DA rats. Ccr2 and Ccl2 levels were not affected after injury in either of the strains (Figure 4D).
Expression levels of pain molecules in spinal cord, L4 DRG, L5 DRG and nerve 7 days after injury (+) and health controls (−) in Aplec and DA rats. IL-1β (A), CGRP L4 and L5 DRG (B), SP L4 and L5 DRG (C), Ccr2, Ccl2, Cx3cr1 and CatS in SC (D). Statistical analysis were done with one-way ANOVA and by Step-down Bonferroni correction for multiple comparison (*p < 0.05; **p < 0.01).
Immune cell infiltration in the spinal cord
Flow cytometry analyses were performed in DA and Aplec, 14 days post-injury to characterize immune cell recruitment to the SC. Populations of microglia, monocyte/macrophages, T-cells and NK cells were analyzed (Figure 5A). As expected, the microglia population greatly outnumbered the other two. There was a tendency for a higher relative proportion of both microglia and T-cells in Aplec compared to DA. In addition, the Aplec strain had a significantly increased proportion of infiltrating activated macrophages (CD45+MHCII+) compared to DA (Figure 5B-F).
Immune cell infiltration to the spinal cord. Flow cytometry analyses performed 14 days after injury in Aplec and DA strains. Representative flow cytometry plots (A). Gated areas represent different cell populations, lymphocytes (a), macrophages (b), microglia (c) T-cells (d) activated infiltrating macrophages (e) and activated microglia (f). Strain differences are presented as percent of total cells and absolute numbers for T-cells (B), infiltrating macrophages (C), activated infiltrating macrophages (D), microglia (E) and activated microglia (F). Statistical analysis were done by Mann–Whitney test (*p < 0.05).
In vitro stimulation of bone marrow derived macrophages
CLRs are expressed by antigen-presenting cells, like macrophages. As we observed an increased infiltration of macrophages to the SC after injury we performed in vitro stimulations of macrophages from both stains to determine possible phenotypic differences in the expression of pro-inflammatory cytokines. Accordingly, bone marrow-derived macrophages (BMMφ) from DA and Aplec rats were stimulated in vitro with TNFα for 24 h. We could detect a significant difference in expression levels of IL-1β, IL-6 and TNFα, where BMMφ cells derived from the Aplec strain expressed higher levels compared to DA (Figure 6A). Interestingly, Dcir1 and Mincle levels were higher in DA strain after stimulation compared to Aplec. Mcl levels were up-regulated after stimulation in both strains but without strain differences (Figure 6B).
Expression levels of pro-inflammatory cytokines, Dcir1, Mincle and Mcl in BMMφ from DA and Aplec following stimulation with TNF- α. The Aplec strain display higher expression of TNF -α , IL-6 and IL-1β (A). Dcir1 and Mincle levels are higher in DA strain after stimulation, Mcl levels are up-regulated in both strains after stimulation but without strain differences (B). One-way ANOVA followed by Bonferroni post-hoc (*p < 0.05; **p < 0.01; ***p < 0.001).
Neuropathic pain-like behavior in the R11R6 congenic rat strain
In order to reproduce our findings, and genetically dissect the Aplec cluster further, a smaller congenic containing only four out of the seven CLRs from the Aplec fragment; Dcir1, Dcar1, Mcl and Mincle, was created. Following SNL the R11R6 developed mechanical hypersensitivity to a similar extent as the Aplec strain and remained sensitive during the entire time of testing (Figure 7). As for Aplec, statistical analysis demonstrated overall differences between the strains, as well as significant differences specifically on day 21, 28 and 35, i.e. the last three time points tested. To assess the cellular phenotype in R11R6 flow cytometry analyse were performed as for the Aplec, however, including only the L4-L6 segment of the cord. As with the Aplec strain the R11R6 had more infiltration of activated macrophages compared to DA. In addition, also the absolute numbers of microglia and infiltrating macrophages were greater in R11R6 compared to DA (Figure 8A-E).
Neuropathic pain-like behavior after peripheral nerve injury in DA and R11R6 strain. One-way ANOVA demonstrates overall differences between the strains (***p < 0.001). Bonferroni post-hoc indicates significant difference between DA and R11R6 on day 21, 28 and 35 (++p < 0.01; +++p < 0.001) Data are expressed as means ± SEM. (12–20 rats per group).
Immune cell regulation to the spinal cord in DA and R11R6 strain 14 days after injury. Flow cytometric analysis using gates as in Figure 5A. Strain differences are presented as percent of total cells and absolute numbers for T-cells (A), infiltrating macrophages (B), activated infiltrating macrophages (C), microglia (D) and activated microglia (E). Statistical analysis were done by Mann–Whitney test (*p < 0.05).
Discussion
In the present study we demonstrate that the Aplec congenic rat displays a nerve injury phenotype distinctly different from DA rats, with continued neuropathic pain-behavior extending well after the DA strain has recovered. The phenotype is associated with increased expression of IL-1β and CatS, as well as increased infiltration of activated macrophages to the SC and a greater response to an inflammatory stimulus of BMMφ in vitro, well in line with the notion of an inflammatory component in neuropathic pain development. The seven CLRs comprised in the cluster are expressed by antigen-presenting cells as well as neutrophils [22, 23] and act as pattern recognition receptors that upon binding of a pathogen or endogenous ligand will shape T -cell responses and modulate the ensuing inflammatory reaction [22, 24, 25]. The Aplec cluster was originally position mapped by comparing the susceptibility of inbred DA rats with that of DA rats carrying alleles derived from the oil-induced arthritis-resistant PVG strain [18]. In a subsequent study the Aplec cluster was found to affect the in vivo and in vitro phenotypes with regard to infectious and inflammatory challenges, further strengthening the notion of effects mediated through the regulation of general macrophage activation status [17]. Interestingly, genetic variability in the corresponding human genes has been associated with susceptibility to anti-citrulline antibody negative rheumatoid arthritis, indicating relevance also for human disease [21].
The role of CLRs has mostly been studied in the context of antigen-presenting cells, in particular dendritic cells, where they have been shown to important for shaping adaptive immune responses [26, 27]. However, detailed knowledge of the molecular function of many CLRs is still lacking and any possible involvement of CLRs in traumatic nerve injuries is entirely unknown. However, in a recent expression quantitative trait loci mapping study we found Dcir3 to be significantly regulated in the SC between DA and PVG after ventral nerve root injury [19]. Interestingly, further testing of the Aplec strain in this injury model revealed an effect on the inflammatory response with more lymphocyte infiltration as well as increased survival of avulsed motoneurons. Here we find a different pattern of regulated genes in the Aplec cluster, with higher expression of Dcir 2 in both SC and L5 DRG of the Aplec strain. In contrast, in the nerve, Dcir 1 and 4 levels were higher in the DA strain. Mincle was not induced by injury, but expression was in general higher in the DA strain. Taken together this suggests that there are complex regulatory differences affecting the expression pattern of several of the CLRs in the fragment underlying the observed phenotypes.
By a large scale breeding effort we were able to identify a sub-congenic with a recombination within the Aplec cluster, isolating four of the seven CLRs in a new fragment; R11R6, containing Dcir1, Dcar1, Mcl and Mincle. Testing of this strain in the SNL model revealed a phenotype almost identical to Aplec, suggesting the underlying genetic variability or variabilities conferring the clinical effect to be localized to this fragment. Of the four genes in the R11R6 fragment Dcar1 could be viewed as a potential candidate since it is nonsense mutated in DA strain [21]. However, the mRNA levels of Dcar1 were barely detectable in all studied tissue arguing against a role for Dcar1. Mcl and Mincle were not induced by injury, but expression of Mincle was higher in the DA strain compared to the Aplec. The last gene, Dcir1, was expressed more highly in the nerve of DA rats and is known to contain an immunoreceptor tyrosine-based inhibitory motif, hence involved in inhibitory signaling [22, 28]. This may imply that DA up-regulate Dcir1 to inhibit activation/secretion of proinflammatory cytokines. In vitro stimulation of BMMφ with TNF-α resulted in higher levels of Dcir1 in DA cells. Given the genetic complexity with possible mutual cross-regulation between the genes in the fragment and differences between different anatomical locations, formal proof of the underlying causative genetic variation or variations may require continued recombinant inbred breeding, an undertaking that could take several years.
We further explored downstream molecular events segregating between the two studied strains. The finding that IL-1β levels were significantly higher in the pain sensitive strain in the SC compared to DA after injury in L5 DRG are in concordance with several studies demonstrating that IL-1β increases neuron excitability and accelerate central sensitization [10, 29]. Expression of IL-1β was also greater in in vitro stimulated BMMφ, suggestin that the Aplec cluster affects expression of this cytokine. As expecte, expression of SP and CGRP was down regulated in both strains after injury, in accordance with pervious knowledge [30–32]. On the contrary we observed an up-regulation of SP and CGRP levels in both strains in the intact L4 DRG, with significantly higher expression of SP in the Aplec strain. Fukuoka et al. observed a similar finding with increased CGRP levels in the contralateral L4 DRG after same type of injury, which may reflect increased activity or sensitivity in intact sensory pain transmission systems, possibly including also inflammatory cytokines [33–36].
Ccl2/Ccr2 and Fractalkine/Cxcr1 are two signaling pathways known to be involved in mediating interaction between injured sensory neurons and microglia [6, 37], in addition Ccl2/Ccr2 signaling has been shown to be important for both monocyte recruitment and pain sensitivity [5, 6]. We could not observe any differences in the expression of Ccl2/Ccr2 in the SC, which could indicate either that the number of recruited macrophages is too low to be detected with this approach or that signaling through this ligand-receptor pair is of less importance in the context studied here. In contrast, we could record an up-regulation of Cx3cr1 in both strains after injury. Interestingly, CatS was up-regulated preferentially in the cord of the pain sensitive Aplec strain. CatS, a proteolytic lysosomal cysteine proteinase, is released by activated microglia in SC and macrophages in the periphery and is responsible for the cleavage of Fractalkine which gives rise to a soluble cleavage product that binds to Cx3cr1 expressing microglia leading to an enhancement of pro-nociceptive mediators [38, 39].
The role of T-cells for the development of neuropathic-pain like behavior is complex, with contrasting effects including both pain-driving and analgesic effects [3, 40–42]. In a previous study of a motor nerve avulsion model we found a greater T -cell infiltration to the SC in Aplec compared to DA [19]. Here we found a tendency for both Aplec and R11R6 rats to have higher numbers of T-cells compared to DA. In contrast, strain differences were evident for infiltration of activated macrophages in Aplec and subsequently confirmed in the R11R6 strain, displaying also increased numbers of microglia as well as macrophages in general.
Previous reports have demonstrated that infiltration of BMMφ to the CNS play a role in the development of neuropathic pain [6]. Hence, the production of cytokines by activated BMMφ cells was examined in vitro using a standard inflammatory stimulus. We could detect that cells derived from the pain sensitive Aplec strain displayed higher expression of TNF-α, IL-1β and IL-6, all of which are known to increase both pain sensitivity and induces the production of each other, which amplifies the inflammatory response [7, 43]. This is in line with a previous study suggesting the Aplec cluster to regulate the general activation status of macrophages [17].
Conclusion
All together our findings support the conclusion that variability in CLRs occurring among inbred rat strains affects inflammatory activation of antigen-presenting cells, with subsequent effects on pain transmitting systems. Importantly, our results, derived from large scale genetic dissection, identified that variability in the four CLR’s in the R11R6 sub-congenic (Dcir1, Dcar1, Mcl and Mincle) is sufficient to cause a significant difference in the clinical effect. Further studies are needed to elucidate the mechanisms more in detail. The fact that this gene cluster was identified by unbiased forward genetics and that genetic variability in human orthologuos have been associated with disease risk encourage studies also in humans.
Materials and methods
Ethics statement
All animal experiments were performed in accordance with the Guidelines of the International Association for the Study of Pain and were approved by the Swedish ethical committee (Stockholm’s North Ethical Committee- Stockholms Norra Djurförsöksetiska nämnd).
Animals
Two congenic rat strains, one containing seven CLR genes denoted antigen-presenting lectin-like receptor complex (Aplec) and the other containing four CLR genes denoted R11R6, as well as the inbred Dark agouti (DA) male rats were used in this study. The congenic Aplec and R11R6 were produced by transferring a gene cluster from Piebald Virol Glaxo (PVG) rats onto Dark-Agouti (DA) rats through repeated backcrossing as previously described [18]. The fragments are on chromosome 4 (222.811-223.365 kb respectively 223.010-223.365 kb) and a schematic map with the gene positions (Gene ID) and markers are depicted in Figure 1 (Ensembl version 73, Rnor 5.0).
All animals were kept under specific pathogen-free and climate-controlled conditions with 12 h light/dark cycles, housed in polystyrene cages containing wood shavings, and fed standard rodent chow and water ad libitum.
Peripheral nerve injury
Rats were subjected to modified spinal nerve ligation model (SNL) [44] under standardized conditions. The animals were deeply anesthetized with 2% isoflurane and lower back skin was shaved and cleaned with 70% ethanol. An incision was made through the skin and paraspinal muscle were separated from the spinous processes at the L5-L6 levels. The fifth lumbar spinal nerve was transected distal to the ganglion. The skin was closed in layers and sutured. 0,25 ml Eusaprim (16 mg/ml sulfametoxazol, 80 mg/ml trimethoprim) (Aspen Europé Gmbh, Bad Oldesloe, Germany) was administrated post surgery, subcutaneously. All rats were sacrificed with CO2 and perfused with PBS containing Heparin (LEO, Pharma AB, Malmö, Sweden). Rats were sacrificed at day 7,14 and 35 after injury.
Behavioral testing
Rat were tested for mechanical hypersensitivity before and on day 3,7,10 and 14 after injury, and then weekly at week 3 and 4. Individual rats were placed in testing chambers with metal mesh floor 10 min before experiments for habituation. A set of calibrated nylon monofilament (Semmes-Weinstein monofilaments, Stoelting, IL) was applied to the glabrous skin of the paws with increasing force until the animal withdrew the limb. Each monofilament was applied 5 times with a few seconds interval and withdrawal threshold was determined when the rat withdrew the paw from at least 3 out of 5 stimulations.
Quantitative real-time PCR (qPCR)
The ipsilateral L5 and adjacent unlesioned L4 dorsal root ganglion (DRG) were identified using a dissection microscope and taken for subsequent analysis. Also the ipsilateral dorsal horn of the spinal cord (SC) (segment L4-L5) and a few millimeter of the nerve proximal to the injury were collected for mRNA quantification at day 7 after SNL. Total RNA was extracted with RNeasy Mini kit (Qiagen) and RNase-Free DNase Set (Qiagen) according to manufacturer’s protocols. cDNA was prepared with 5x iScript reaction mix (Bio-Rad) with 5 μl total RNA. Amplifications were conducted using Bio-Rad SYBR green according to manufacturer’s instructions and plates were run in Bio-Rad CFX optical system (Bio-Rad). Primer specificity was assessed by determining amplicon size using gel electrophoresis and melt curve analysis of each reaction indicating a single peak. The targets analyzed and their primer sequences are listed in Table 1. Normalized expressions were calculated in Bio Rad CFX manager v2.0 (Bio-Rad) using hprt and gapdh as house-keeping genes.
Flow cytometry
At day 14 after injury animals were scarified with CO2 and perfused through the ascending aorta with ice-cold PBS supplemented with heparin (LEO Pharma AB, Malmö, Sweden). The spinal cords (n = 5-7 rats/strain) were removed and homogenized with a glass tissue grinder in a 50% Percoll solution (Sigma-Aldrich, Stockholm, Sweden). A density gradient was made consisting of the following layers: a top layer of 30% Percoll (20 ml), a middle layer with the homogenized tissue in 50% Percoll (20 ml) and a bottom layer of 63% Percoll (7 ml). All Percoll solutions were made fresh by diluting Percoll in 10xHBSS (Hank’s Balanced Salt Solution, Gibco), supplemented with 0.1% BSA and 0.1% glucose. After centrifugation at 1000 g at 7°C for 30 min, cells below the myelin layer were collected, washed with PBS containing 0.5% FBS and 2 mM EDTA and stained with the following antibodies: CD3-FITC, MHCII-PerCP, CD45-APC and CD11b-APC-Cy7 (eBioscience). Samples were run in Gallios flow cytometer (Beckman Coulter, Brea, USA) and analysis of acquired cells was performed with Kaluza v1.1 (Beckman Coulter). In the first experiment done on Aplec and DA rats the whole spinal cord was taken for analysis whereas for the R11R6 and DA experiment only the lumbar segment of interest, L4-L6 was taken for analysis.
Bone marrow-derived macrophages culture
Bone marrow-derived macrophages were cultured as described previously [45] from naive DA and Aplec rats. In brief, femurs were dissected and femoral bone-marrow cells were collected by flushing through medium with a 21-gauge needle. Single-cell suspensions were prepared and re-suspended in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) supplemented with 20% heat-inactivated FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine and 20% M-CSF conditioned L929 cell line supernatant. Bone marrow cells were cultured in 175 cm2 cell culture flasks and incubated at 37°C and 5% C02 in a humidified incubator for 10 days; medium was changed after 4 and 6 days with M-CSF conditioned medium, and after 10 days with complete medium (DMEM + FBS) without M-CSF conditioning. Cells were harvested using EDTA (Sigma) at a concentration of 0.5 mM and seeded in 24-well plates (4 × 105 cells/well).
The cells were then left un-stimulated, or stimulated with TNF-α (20 ng/ml) for 24 h and then taken for analysis with RT-PCR.
Statistical analysis
Statistical analyses were conducted using Graphpad Prism (5.0). For behavioral analysis one-way ANOVA data analysis were performed for overall differences (***p < 0.001) followed by Bonferroni post-hoc for individual time points (++p < 0.01; +++p < 0.001). Data are expressed as means ± standard error of the mean. For in vivo RT-PCR analysis one-way ANOVA was done followed by Step-down Bonferroni correction for multiple comparisons. The in vitro RT-PCR analysis were done by one way ANOVA followed by Bonferroni post-hoc (*p < 0.05; **p < 0.01; ***p < 0.001). The flow cytometry studies were analysed with Mann–Whitney test (*p < 0.05).
References
Watkins LR, Maier SF: Beyond neurons: evidence that immune and glial cells contribute to pathological pain states. Physiol Rev 2002, 82: 981–1011.
DeLeo JA, Yezierski RP: The role of neuroinflammation and neuroimmune activation in persistent pain. Pain 2001, 90: 1–6. 10.1016/S0304-3959(00)00490-5
Costigan M, Moss A, Latremoliere A, Johnston C, Verma-Gandhu M, Herbert TA, Barrett L, Brenner GJ, Vardeh D, Woolf CJ, Fitzgerald M: T-cell infiltration and signaling in the adult dorsal spinal cord is a major contributor to neuropathic pain-like hypersensitivity. J Neurosci 2009, 29: 14415–14422. 10.1523/JNEUROSCI.4569-09.2009
Sweitzer SM, Hickey WF, Rutkowski MD, Pahl JL, DeLeo JA: Focal peripheral nerve injury induces leukocyte trafficking into the central nervous system: potential relationship to neuropathic pain. Pain 2002, 100: 163–170. 10.1016/S0304-3959(02)00257-9
Mildner A, Schmidt H, Nitsche M, Merkler D, Hanisch UK, Mack M, Heikenwalder M, Bruck W, Priller J, Prinz M: Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat Neurosci 2007, 10: 1544–1553. 10.1038/nn2015
Zhang J, Shi XQ, Echeverry S, Mogil JS, De Koninck Y, Rivest S: Expression of CCR2 in both resident and bone marrow-derived microglia plays a critical role in neuropathic pain. J Neurosci 2007, 27: 12396–12406. 10.1523/JNEUROSCI.3016-07.2007
Sommer C, Kress M: Recent findings on how proinflammatory cytokines cause pain: peripheral mechanisms in inflammatory and neuropathic hyperalgesia. Neurosci Lett 2004, 361: 184–187. 10.1016/j.neulet.2003.12.007
Sommer C, Schafers M: Painful mononeuropathy in C57BL/Wld mice with delayed wallerian degeneration: differential effects of cytokine production and nerve regeneration on thermal and mechanical hypersensitivity. Brain Res 1998, 784: 154–162. 10.1016/S0006-8993(97)01327-9
Austin PJ, Moalem-Taylor G: The neuro-immune balance in neuropathic pain: involvement of inflammatory immune cells, immune-like glial cells and cytokines. J Neuroimmunol 2010, 229: 26–50. 10.1016/j.jneuroim.2010.08.013
Marchand F, Perretti M, McMahon SB: Role of the immune system in chronic pain. Nat Rev Neurosci 2005, 6: 521–532.
Wiesenfeld-Hallin Z, Hao JX, Xu XJ, Aldskogius H, Seiger A: Genetic factors influence the development of mechanical hypersensitivity, motor deficits and morphological damage after transient spinal cord ischemia in the rat. Pain 1993, 55: 235–241. 10.1016/0304-3959(93)90152-F
Dominguez CA, Strom M, Gao T, Zhang L, Olsson T, Wiesenfeld-Hallin Z, Xu XJ, Piehl F: Genetic and sex influence on neuropathic pain-like behaviour after spinal cord injury in the rat. Eur J Pain 2012, 16: 1368–1377. 10.1002/j.1532-2149.2012.00144.x
Mogil JS, Wilson SG, Bon K, Lee SE, Chung K, Raber P, Pieper JO, Hain HS, Belknap JK, Hubert L, Elmer GI, Chung JM, Devor M: Heritability of nociception I: responses of 11 inbred mouse strains on 12 measures of nociception. Pain 1999, 80: 67–82. 10.1016/S0304-3959(98)00197-3
Devor M, Gilad A, Arbilly M, Yakir B, Raber P, Pisante A, Darvasi A: pain1: a neuropathic pain QTL on mouse chromosome 15 in a C3HxC58 backcross. Pain 2005, 116: 289–293. 10.1016/j.pain.2005.04.023
Dominguez CA, Lidman O, Hao JX, Diez M, Tuncel J, Olsson T, Wiesenfeld-Hallin Z, Piehl F, Xu XJ: Genetic analysis of neuropathic pain-like behavior following peripheral nerve injury suggests a role of the major histocompatibility complex in development of allodynia. Pain 2008, 136: 313–319. 10.1016/j.pain.2007.07.009
Dominguez CA, Kalliomaki M, Gunnarsson U, Moen A, Sandblom G, Kockum I, Lavant E, Olsson T, Nyberg F, Rygh LJ, Roe C, Gjerstad J, Gordh T, Piehl F: The DQB1 *03:02 HLA haplotype is associated with increased risk of chronic pain after inguinal hernia surgery and lumbar disc herniation. Pain 2013, 154: 427–433. 10.1016/j.pain.2012.12.003
Guo JP, Verdrengh M, Tarkowski A, Lange S, Jennische E, Lorentzen JC, Harris RA: The rat antigen-presenting lectin-like receptor complex influences innate immunity and development of infectious diseases. Genes Immun 2009, 10: 227–236. 10.1038/gene.2009.4
Flornes LM, Bryceson YT, Spurkland A, Lorentzen JC, Dissen E, Fossum S: Identification of lectin-like receptors expressed by antigen presenting cells and neutrophils and their mapping to a novel gene complex. Immunogenetics 2004, 56: 506–517. 10.1007/s00251-004-0714-x
Lindblom RP, Aeinehband S, Parsa R, Strom M, Al Nimer F, Zhang XM, Dominguez CA, Flytzani S, Diez M, Piehl F: Genetic variability in the rat Aplec C-type lectin gene cluster regulates lymphocyte trafficking and motor neuron survival after traumatic nerve root injury. J Neuroinflammation 2013, 10: 60. 10.1186/1742-2094-10-60
Guo JP, Backdahl L, Marta M, Mathsson L, Ronnelid J, Lorentzen JC: Profound and paradoxical impact on arthritis and autoimmunity of the rat antigen-presenting lectin-like receptor complex. Arthritis Rheum 2008, 58: 1343–1353. 10.1002/art.23434
Lorentzen JC, Flornes L, Eklow C, Backdahl L, Ribbhammar U, Guo JP, Smolnikova M, Dissen E, Seddighzadeh M, Brookes AJ, Alfredsson L, Klareskog L, Padyukov L, Fossum S: Association of arthritis with a gene complex encoding C-type lectin-like receptors. Arthritis Rheum 2007, 56: 2620–2632. 10.1002/art.22813
Geijtenbeek TB, Gringhuis SI: Signalling through C-type lectin receptors: shaping immune responses. Nat Rev Immunol 2009, 9: 465–479. 10.1038/nri2569
Lobato-Pascual A, Saether PC, Dahle MK, Gaustad P, Dissen E, Fossum S, Daws MR: Rat macrophage C-type lectin is an activating receptor expressed by phagocytic cells. PLoS One 2013, 8: e57406. 10.1371/journal.pone.0057406
Desel C, Werninghaus K, Ritter M, Jozefowski K, Wenzel J, Russkamp N, Schleicher U, Christensen D, Wirtz S, Kirschning C, Agger EM, Prazeres da Costa C, Lang R: The Mincle-activating adjuvant TDB induces MyD88-dependent Th1 and Th17 responses through IL-1R signaling. PLoS One 2013, 8: e53531. 10.1371/journal.pone.0053531
Meyer-Wentrup F, Cambi A, Joosten B, Looman MW, de Vries IJ, Figdor CG, Adema GJ: DCIR is endocytosed into human dendritic cells and inhibits TLR8-mediated cytokine production. J Leukoc Biol 2009, 85: 518–525.
Kaden SA, Kurig S, Vasters K, Hofmann K, Zaenker KS, Schmitz J, Winkels G: Enhanced dendritic cell-induced immune responses mediated by the novel C-type lectin receptor mDCAR1. J Immunol 2009, 183: 5069–5078. 10.4049/jimmunol.0900908
Meyer-Wentrup F, Benitez-Ribas D, Tacken PJ, Punt CJ, Figdor CG, de Vries IJ, Adema GJ: Targeting DCIR on human plasmacytoid dendritic cells results in antigen presentation and inhibits IFN-alpha production. Blood 2008, 111: 4245–4253. 10.1182/blood-2007-03-081398
Fujikado N, Saijo S, Yonezawa T, Shimamori K, Ishii A, Sugai S, Kotaki H, Sudo K, Nose M, Iwakura Y: Dcir deficiency causes development of autoimmune diseases in mice due to excess expansion of dendritic cells. Nat Med 2008, 14: 176–180. 10.1038/nm1697
Fukuoka H, Kawatani M, Hisamitsu T, Takeshige C: Cutaneous hyperalgesia induced by peripheral injection of interleukin-1 beta in the rat. Brain Res 1994, 657: 133–140. 10.1016/0006-8993(94)90960-1
Hokfelt T, Zhang X, Wiesenfeld-Hallin Z: Messenger plasticity in primary sensory neurons following axotomy and its functional implications. Trends Neurosci 1994, 17: 22–30. 10.1016/0166-2236(94)90031-0
Noguchi K, Senba E, Morita Y, Sato M, Tohyama M: Prepro-VIP and preprotachykinin mRNAs in the rat dorsal root ganglion cells following peripheral axotomy. Brain Res Mol Brain Res 1989, 6: 327–330. 10.1016/0169-328X(89)90077-6
Doughty SE, Atkinson ME, Shehab SA: A quantitative study of neuropeptide immunoreactive cell bodies of primary afferent sensory neurons following rat sciatic nerve peripheral axotomy. Regul Pept 1991, 35: 59–72. 10.1016/0167-0115(91)90254-E
Ma W, Quirion R: Increased calcitonin gene-related peptide in neuroma and invading macrophages is involved in the up-regulation of interleukin-6 and thermal hyperalgesia in a rat model of mononeuropathy. J Neurochem 2006, 98: 180–192. 10.1111/j.1471-4159.2006.03856.x
Fukuoka T, Tokunaga A, Kondo E, Miki K, Tachibana T, Noguchi K: Change in mRNAs for neuropeptides and the GABA(A) receptor in dorsal root ganglion neurons in a rat experimental neuropathic pain model. Pain 1998, 78: 13–26. 10.1016/S0304-3959(98)00111-0
Cuesta MC, Quintero L, Pons H, Suarez-Roca H: Substance P and calcitonin gene-related peptide increase IL-1 beta, IL-6 and TNF alpha secretion from human peripheral blood mononuclear cells. Neurochem Int 2002, 40: 301–306. 10.1016/S0197-0186(01)00094-8
Schafers M, Lee DH, Brors D, Yaksh TL, Sorkin LS: Increased sensitivity of injured and adjacent uninjured rat primary sensory neurons to exogenous tumor necrosis factor-alpha after spinal nerve ligation. J Neurosci 2003, 23: 3028–3038.
Abbadie C, Bhangoo S, De Koninck Y, Malcangio M, Melik-Parsadaniantz S, White FA: Chemokines and pain mechanisms. Brain Res Rev 2009, 60: 125–134. 10.1016/j.brainresrev.2008.12.002
Clark AK, Yip PK, Grist J, Gentry C, Staniland AA, Marchand F, Dehvari M, Wotherspoon G, Winter J, Ullah J, Bevan S, Malcangio M: Inhibition of spinal microglial cathepsin S for the reversal of neuropathic pain. Proc Natl Acad Sci U S A 2007, 104: 10655–10660. 10.1073/pnas.0610811104
Barclay J, Clark AK, Ganju P, Gentry C, Patel S, Wotherspoon G, Buxton F, Song C, Ullah J, Winter J, Fox A, Bevan S, Malcangio M: Role of the cysteine protease cathepsin S in neuropathic hyperalgesia. Pain 2007, 130: 225–234. 10.1016/j.pain.2006.11.017
Moalem G, Xu K, Yu L: T lymphocytes play a role in neuropathic pain following peripheral nerve injury in rats. Neuroscience 2004, 129: 767–777. 10.1016/j.neuroscience.2004.08.035
Cui JG, Holmin S, Mathiesen T, Meyerson BA, Linderoth B: Possible role of inflammatory mediators in tactile hypersensitivity in rat models of mononeuropathy. Pain 2000, 88: 239–248. 10.1016/S0304-3959(00)00331-6
Labuz D, Schreiter A, Schmidt Y, Brack A, Machelska H: T lymphocytes containing beta-endorphin ameliorate mechanical hypersensitivity following nerve injury. Brain Behav Immun 2010, 24: 1045–1053. 10.1016/j.bbi.2010.04.001
Watkins LR, Nguyen KT, Lee JE, Maier SF: Dynamic regulation of proinflammatory cytokines. Adv Exp Med Biol 1999, 461: 153–178. 10.1007/978-0-585-37970-8_10
Kim SH, Chung JM: An experimental model for peripheral neuropathy produced by segmental spinal nerve ligation in the rat. Pain 1992, 50: 355–363. 10.1016/0304-3959(92)90041-9
Weischenfeldt J, Porse B: Bone Marrow-Derived Macrophages (BMM): isolation and applications. CSH Protoc 2008, 2008: pdb prot5080.
Acknowledgements
We would like to thank Dr Jian Ping Guo for providing the initial Aplec breeding pairs. We would also like to thank Brinda Acharjee for help with genotyping the congenics. This study was supported by the 7th Framework Program of the European Union, EURATrans, HEALTH-F4-2010-241504, by the Swedish Research Council and the Swedish Brain Foundation and the Swedish Association of Persons with Neurological Disabilities. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CD designed the study, performed and planed all experimental work, analyzed the results and wrote the manuscript. XMZ, FAN and AOGC performed and analyzed the flow cytometry analysis. KC assisted in the experimental work and performed expression analyses. RL assisted in experimental planning and analyzed results. FP designed the study, analyzed the data and wrote the first manuscript draft. All authors have read and approved the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Dominguez, C.A., Carlström, K.E., Zhang, XM. et al. Variability in C-type lectin receptors regulates neuropathic pain-like behavior after peripheral nerve injury. Mol Pain 10, 78 (2014). https://doi.org/10.1186/1744-8069-10-78
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1744-8069-10-78