Abstract
Background
Cancerous inhibitor of protein phosphatase 2A (CIP2A) is an oncoprotein that acts as a prognostic marker for several human malignancies. In this study, we investigated the clinical significance of CIP2A and its function in nasopharyngeal carcinoma (NPC).
Methods
Quantitative RT-PCR, western blot, and immunohistochemistry analyses were used to quantify CIP2A expression in NPC cell lines and clinical samples. Kaplan-Meier curves were used to estimate the association between CIP2A expression and patient survival. The functional role of CIP2A in NPC cell lines was evaluated by small interfering RNA-mediated depletion of the protein followed by analyses of cell proliferation and xenograft growth.
Results
CIP2A levels were upregulated in NPC cell lines and clinical samples at both the mRNA and protein levels (P < 0.01). Patients with high CIP2A expression had poorer overall survival (HR, 1.98; 95% CI, 1.16-3.34; P = 0.01) and poorer disease-free survival (HR, 1.68; 95% CI, 1.07-2.62; P = 0.02) rates than patients with low CIP2A expression. In addition, CIP2A expression status was an independent prognostic indicator for NPC patients. The depletion of CIP2A expression inhibited c-Myc protein expression in NPC cell lines, suppressed cell viability, colony formation, and anchorage-independent growth in vitro, and inhibited xenograft tumor growth in vivo.
Conclusions
Our data demonstrate that high CIP2A expression in patients was associated with poor survival in NPC, and depletion of CIP2A expression inhibited NPC cell proliferation and tumor growth. Thus, these results warrant further investigation of CIP2A as a novel therapeutic target for the treatment of NPC.
Similar content being viewed by others
Introduction
Nasopharyngeal carcinoma (NPC) is an epithelial malignancy of the nasopharynx, and global statistics obtained for different world regions reveal that its distribution is extremely unbalanced, with the highest incidence rates occurring in Southern China [1, 2]. According to data from the International Agency for Research on Cancer, there were an estimated 84,000 cases of NPC and 51,600 NPC-related deaths in 2008 [3]. Although the increased prevalence of intensity-modulated radiation therapy with concurrent chemoradiation therapy has improved the local and regional control of NPC for patients with locoregionally advanced disease, their prognosis is still poor due to recurrence and/or distant metastasis [4, 5]. Thus, a better understanding of the underlying molecular mechanisms involved in NPC pathogenesis and progression is essential for the development of novel therapeutic strategies to treat patients with NPC.
Cancerous inhibitor of protein phosphatase 2A (CIP2A), also known as KIAA1524 and p90, is a recently identified human oncoprotein that inhibits the degradation of c-MYC by inhibiting the protein phosphatase 2A (PP2A)-mediated dephosphorylation of MYC at serine 62 [6–8]. In addition, CIP2A and MYC appear to be regulated by a positive feedback loop that promotes the expression of both proteins [7]. Originally identified as an oncoprotein, CIP2A contributes to the immortalization and malignant transformation of human cells [6], and the depletion of CIP2A decreased cell viability and anchorage-independent growth in a variety of human malignancies [6–11]. More importantly, CIP2A was recently found to be overexpressed at a high frequency in most types of cancer and may serve as a prognostic predictor [12–20]. However, the clinical significance and biological function of CIP2A in NPC has not been thoroughly investigated to date.
In the present study, we examined both the mRNA and protein expression levels of CIP2A in NPC cell lines and tissue samples and further analyzed the clinical significance of CIP2A in a cohort of NPC patients. In addition, we explored the potential role of CIP2A in NPC cell proliferation and tumor growth, which could help to better understand the pathology of NPC and may further provide a novel therapeutic target for the treatment of NPC patients.
Results
Expression of CIP2A in NPC cells and tissues
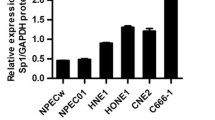
Quantitative RT-PCR and western blot analyses were used to determine the levels of CIP2A mRNA and protein in NPC cell lines and the normal nasopharyngeal epithelial cell line NP69. CIP2A was significantly upregulated in all six NPC cell lines when compared to the NP69 cells at both the mRNA and protein levels (Figure 1A-B). Furthermore, we detected CIP2A mRNA expression in 18 freshly frozen NPC tissues and 14 normal nasopharyngeal epithelial tissues and found that CIP2A mRNA levels were considerably higher in NPC tissues (Figure 1C). Similarly, CIP2A protein was also increased in NPC tissues when compared to normal nasopharyngeal epithelial tissues (Figure 1D). These results suggest that CIP2A is upregulated in NPC.
Expression levels of CIP2A in NPC cell lines and clinical samples. (A-B) Expression levels of CIP2A mRNA (A) and protein (B) in NP69 and NPC cell lines. (C-D) Expression levels of CIP2A mRNA (C) and protein (D) in NPC tissues and normal nasopharyngeal epithelial tissues. GAPDH was used as the endogenous control. Data are presented as the mean ± SD, and P values were calculated with Student’s t-test.
CIP2A expression and the clinical variables of NPC patients
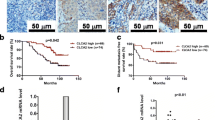
We then analyzed CIP2A protein expression levels in a set of 280 paraffin-embedded NPC tissue samples using immunohistochemistry. Representative staining of CIP2A in NPC tissue is shown in Figure 2A-H, and positive staining of CIP2A was mainly observed in the cytoplasm. The presence of CIP2A protein was detected in 254 of the 280 (90.3%) cancer samples analyzed, and CIP2A protein expression was highly expressed in 184 of the 280 (65.7%) NPC patients examined. Furthermore, patients with high CIP2A expression exhibited a significant association with T stage (P = 0.04), TNM stage (P < 0.01), distant metastasis (P < 0.01), and patient death (P = 0.01). There were no significant associations between CIP2A expression and patient age, sex, WHO type, VCA-IgA, EA-IgA, N stage, or locoregional failure (Table 1).
Expression levels of CIP2A and survival of NPC patients. (A-H) Dicer1 protein expression is mainly localized to the cytoplasm. (A, E) Negative staining (A: 200×; E: 400×); (B, F) Weak staining: light yellow (B: 200×; F: 400×); (C, G) Moderate staining: yellow brown (C: 200×; G: 400×). (D, H) Strong staining: brown (D: 200×; H: 400×). (I-J) Patients with high CIP2A expression had poorer overall survival (I) and poorer disease-free survival (J) rates than patients with low CIP2A expression.
CIP2A expression and survival of NPC patients
Kaplan-Meier analysis and the log-rank test were used to calculate the effects of CIP2A on survival, and the results indicated that patients with high CIP2A expression were significantly associated with poorer overall and disease-free survival rates than patients with low CIP2A expression (all P < 0.01, Figure 2I-J). The cumulative 5-year survival rate was 86.5% (95% CI, 79.7-93.3) in the low-CIP2A-expression group, whereas it was only 74.5% (95% CI, 68.2-80.8) in the high-CIP2A-expression group. CIP2A expression, TNM stage, sex, age, WHO type, and EBV seromarkers were analyzed using univariate and multivariate Cox regression analyses. Univariate analyses indicated that patients with high CIP2A expression and advanced disease stages exhibited worse outcomes than those with low CIP2A expression (Table 2). Multivariate analyses revealed that CIP2A expression and TNM stage were independent prognostic indicators in NPC patients (all P < 0.05, Table 2).
Effects of CIP2A depletion on MYC expression and cell proliferation
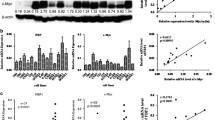
CIP2A protein expression was remarkably inhibited in CNE-2 and SUNE-1 cells treated with siRNA specifically directed against CIP2A when compared to those treated with scrambled control siRNA (Figure 3A-B). More importantly, depletion of CIP2A by siRNA suppressed the MYC protein expression in both CNE-2 and SUNE-1 cells (Figure 3A-B). We also studied the effects of CIP2A depletion on cell viability and proliferation ability using MTT assays and colony formation assays. CNE-2 and SUNE-1 cells transfected with siCIP2A displayed significant growth inhibition compared to those transfected with scrambled control siRNA (P < 0.05, Figure 3C). Moreover, cells transfected with siCIP2A exhibited fewer and smaller colonies compared to the controls (P < 0.05, Figure 3D).
Effects of CIP2A depletion on MYC expression and NPC cell proliferation in vitro. (A-B) Effects of CIP2A siRNA on CIP2A and MYC protein expression in CNE-2 and SUNE-1 cells detected by western blot analysis (A) and immunofluorescence staining (B); (C-E) Effects of CIP2A siRNA on the cell viability (C), proliferation (D), and anchorage-independent growth (E) of CNE-2 and SUNE-1 cells. Data are presented as the mean ± SD. *P < 0.05 compared to the control using Student’s t-test.
Effects of CIP2A depletion on tumor growth
The ability of cells to grow and form colonies on soft agar is a hallmark characteristic of malignantly transformed cells. To study the effects of CIP2A on the malignant growth of NPC cells, we transfected siCIP2A or scrambled control siRNA into CNE-2 and SUNE-1 cells and found that CIP2A depletion significantly suppressed the anchorage-independent growth of both CNE-2 and SUNE-1 cells (all P < 0.05, Figure 3E). To further explore whether CIP2A was required for NPC tumor growth in vivo, we conducted xenograft tumor model assays by subcutaneously injecting SUNE-1 cells stably expressing shCIP2A or scrambled control siRNA into the dorsal flank of several mice. CIP2A depletion resulted in a significant reduction in tumor growth (P < 0.01, Figure 4A-B). The average tumor weight was also significantly decreased in the CIP2A depletion group compared to the scrambled control siRNA-treated group (0.31 ± 0.11 g vs. 0.54 ± 0.24 g; P < 0.01, Figure 4C-D).
Effects of CIP2A depletion on NPC xenograft tumor growth in vivo. (A) SUNE-1 cells stably expressing shCIP2A or scrambled control siRNA were subcutaneously injected into nude mice. SUNE-1 cells stably expressing shCIP2A had smaller tumors than scrambled controls after four weeks. (B) The growth curves of tumor volumes. (C) Representative picture of xenograft tumors. (D) Tumor weight. Data are presented as the mean ± SD. *P < 0.05 compared to the control using Student’s t-test.
Discussion
In this study, CIP2A was upregulated in both NPC cell lines and clinical samples, and those NPC patients with high CIP2A expression exhibited the poorest survival rates. Furthermore, silencing CIP2A expression influenced MYC protein expression and further suppressed NPC cell proliferation and tumor growth. Our results demonstrate that the overexpression of CIP2A plays important roles in the development and progression of NPC.
Reversible protein phosphorylation is one of the most important biological mechanisms for signal transduction, which is tightly regulated by protein kinases and phosphatases to maintain the balance of the protein’s phosphorylation status and control its biological functions. However, there is considerable evidence indicating that the perturbation of this balance, including the activation of protein kinases and inhibition of phosphatases, contributes to the origin and pathogenesis of several human diseases, including cancer [21]. Protein phosphatase 2A (PP2A) is one important type of serine/threonine phosphatase; PP2A is inhibited in human cancers and functions as a tumor suppressor. Furthermore, the inhibition of PP2A activity has been found to result in the immortalization and malignant transformation of human cells [22, 23]. Interestingly, CIP2A has recently been identified as an endogenous PP2A inhibitor in human cancer cells using the tandem affinity purification method. In addition, CIP2A inhibition has been found to enhance the catalytic phosphatase activity of the PP2A complex in several types of human malignancies [6, 24]. Furthermore, CIP2A also exhibits the ability to transform human immortalized cells [6]; these results expand the general understanding of the mechanisms that are critical for cancer development and progression.
CIP2A was previously demonstrated to be a human oncoprotein due to its ability to transform human immortalized cells. Recently, CIP2A was found to be overexpressed at high frequencies in several types of human cancers [6–11]. More importantly, several studies reported that CIP2A could serve as a prognostic indicator for various solid and hematological tumors, including non-small cell lung cancer [12], colon cancer [13], breast cancer [14], ovarian cancer [15], renal cancer [16], tongue cancer [17], esophageal adenocarcinoma [18], bladder cancer [19], and chronic myeloid leukemia [20]. In the present study, CIP2A was significantly overexpressed in NPC cell lines and clinical specimens at both the mRNA and protein levels. Strikingly, NPC patients with high CIP2A protein expression had poorer overall and disease-free survival rates than those with low CIP2A protein expression. Multivariate Cox regression analysis demonstrated that low CIP2A protein expression was an independent prognostic indicator in patients with NPC. These results suggest that CIP2A expression status can serve as a valuable prognostic biomarker to stratify NPC patients into different risk groups and further guide individual therapy choices for patients with NPC.
In addition to its biological significance in the promotion of malignant transformation of human cells, CIP2A also plays important roles in carcinogenesis and the progression of human cancers. Several recent studies have reported that silencing CIP2A decreases cell viability and suppresses anchorage-independent growth in several types of human cancer cells [6–11]. It also promotes progenitor cell self-renewal and protects cancer cells from therapy-induced apoptosis or the induction of senescence [25, 26]. A recent study demonstrated that CIP2A can regulate the cell cycle by targeting PLK1 [27]. More importantly, recent studies have also demonstrated that the depletion of CIP2A via siRNAs inhibits xenograft tumor growth [6, 9]. In our present study, we also depleted CIP2A expression via siRNA to better understand the function of CIP2A in NPC. Inhibition of CIP2A expression significantly inhibited NPC cell viability and proliferation in vitro. Furthermore, silencing CIP2A suppressed xenograft tumor growth in vivo. Taken together, these results demonstrate that the dysregulation of CIP2A may contribute to the development and progression of NPC.
In addition, the depletion of CIP2A expression via siRNA suppressed MYC protein expression in NPC cell lines. MYC is one of the most-studied oncogenes, and it is involved in several malignant cellular processes. CIP2A can inhibit the degradation of MYC and therefore enhance its oncogenic activities by inhibiting the PP2A-mediated dephosphorylation of MYC at serine 62 [6–8]. CIP2A and MYC are regulated by a positive feedback loop that promotes the expression of both proteins [7]. Furthermore, the mechanisms of CIP2A activation and overexpression in cancer cells has been investigated by several other studies in which E2F1, ETS1, and ATF2 were found to directly bind to the CIP2A promoter and further stimulate CIP2A transcription [28–30]. Based on the functions and mechanisms of CIP2A activation in human cancers, the therapeutic targeting of CIP2A could facilitate a novel strategy for cancer therapy, including the use of CIP2A small RNA interference technology or the development of small molecules that target the CIP2A-PP2A interaction [6, 31]. In addition, another alternative strategy to inhibit CIP2A activity is to target the signaling mechanisms that drive high CIP2A expression, such as the use of MYC [7], EGFR [29], and MEK inhibitors [29].
Conclusions
In conclusion, the present study indicated that CIP2A overexpression was associated with poor survival in patients with NPC, and the depletion of CIP2A expression could inhibit cell viability and growth by promoting the stability of the CIP2A protein. Our findings provide new insights into the molecular mechanisms involved in the regulation of NPC progression and provide novel therapeutic targets and strategies for the treatment of NPC patients.
Materials and methods
Cell culture
Human NPC cell lines (CNE-1, CNE-2, SUNE-1, C666-1, HNE1, and HONE1) were grown in RPMI-1640 (Invitrogen) medium supplemented with 10% fetal bovine serum (Gibco). The immortalized nasopharyngeal epithelial cell line NP69 was cultured in keratinocyte/serum-free medium (Invitrogen) supplemented with bovine pituitary extract. The 293FT cell line was maintained in DMEM (Invitrogen) supplemented with 10% fetal bovine serum.
Clinical specimens
Eighteen freshly frozen NPC specimens and fourteen normal nasopharyngeal epithelium samples were obtained from Sun Yat-sen University Cancer Center. In addition, we collected 280 paraffin-embedded NPC specimens from our hospital between January 2003 and February 2006. None of the patients received any anti-tumor therapy prior to the biopsy sample collection. The clinical features of all patients are provided in Table 1. TNM staging was performed according to the 7th Edition of the AJCC/UICC Cancer Staging Manual. All patients were treated with conventional two-dimensional radiotherapy, and patients with stage III-IV disease also received platinum-based concurrent chemotherapy [32]. The median follow-up time was 63.6 months (range: 5.2-91.87). This study was approved by the Institutional Ethical Review Board of Sun Yat-sen University Cancer Center, and written informed consent was obtained from each patient.
RNA extraction, reverse transcription, and quantitative PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) and reverse-transcribed using M-MLV reverse transcriptase (Promega) and random primers (Promega). Quantitative PCR reactions using a Platinum SYBR Green qPCR SuperMix-UDG reagent (Invitrogen) were performed with a Bio-Rad CFX96 sequence detection system (Bio-Rad Laboratories Inc.). The following primers were used for CIP2A: 5′-CCATATGCTCACTCAGATGATGT-3′ (forward) and 5′-GTGTATCATCTCCACAGAGAGTT-3′ (reverse). Primers for GAPDH were as follows: 5′-CTCCTCCTGTTCGACAGTCAGC-3′ (forward) and 5′-CCCAATACGACCAAATCCGTT -3′ (reverse). Reactions containing either no template or no reverse transcriptase were used as negative controls. GAPDH was used as the normalization control, and the relative expression levels were calculated by the 2-ΔΔCT method [33].
Western blot analysis
Total protein was extracted with sample buffer (62.5 mmol/L Tris–HCl, pH 6.8, 2% SDS, 10% glycerol, and 5% 2-β-mercaptoethanol), and its concentration was quantified using the Pierce® BCA Protein Assay Kit (Thermo). Total protein was subsequently separated on 10% SDS-PAGE gels and transferred onto polyvinylidene fluoride membranes (Millipore). The membranes were blocked with 5% skim milk and incubated with primary antibodies recognizing CIP2A (1:1,000; Abcam) and MYC (1:5,000; Epitomics), followed by incubation with anti-mouse or rabbit IgG secondary antibodies (1:5,000; Sigma). Bands were detected by enhanced chemiluminescence, and GAPDH levels (1:5,000; Epitomics antibody) served as the loading control.
Immunohistochemistry
Sections obtained from 280 paraffin-embedded NPC specimens were tested for CIP2A expression by immunohistochemical staining, as previously described [34, 35]. Briefly, samples were deparaffinized and rehydrated, and the endogenous peroxidase activity was quenched. Antigen retrieval was performed, and the sections were blocked with bovine serum albumin and subsequently incubated with an anti-CIP2A antibody (1:200; Abcam). Sections were washed and subsequently incubated with a biotinylated secondary antibody bound to a streptavidin-horseradish peroxidase complex and visualized with 3,3-diaminobenzidine. All sections were scored by two independent pathologists, and the staining index was calculated as the product of the staining intensity (1, no staining; 2, weak staining; 3, moderate staining; 4, strong staining) and the proportion of positive cells (1, <10%; 2, 10-35%; 3, 35-70%; 4, >70%).
Small interfering RNA transfection and the generation of stably transfected cell lines
Double-stranded small interfering RNA targeting CIP2A (50 nM, GenePharma; siCIP2A 1 5′-CUGUGGUUGUGUUUGCACUTT-3′; siCIP2A 2 5′-ACCAUUGAUAUCCUUAGAATT -3′) or scrambled control siRNA (5′-UAACAAUGAGAGCACGGCTT-3′) were transfected into CNE2 and SUNE1 cell lines using Lipofectamine 2000 reagent (Invitrogen). The CIP2A short hairpin RNA (shRNA) was synthesized and cloned into a pSUPERretro-puromycin plasmid using Bgl II and EcoR I enzymes (New England Biolabs). The pSUPERretro-shCIP2A plasmid or empty vector was co-transfected into 293FT cells along with the retroviral packaging vector PIK. After transfection, the supernatants were harvested and used to infect SUNE1 cells, and the stably transfected cells were selected with puromycin and validated by western blot analysis.
Immunofluorescence staining
CNE-2 and SUNE-1 cells were grown on coverslips (Fisher Scientific). After 24 h, cells were incubated with primary antibodies against CIP2A (1:200; Abcam) and MYC (1:200; Epitomics), and subsequently incubated with Alexa Fluor® 488 or 594 goat anti-mouse or anti-rabbit IgG antibodies (Life Technologies). The coverslips were counterstained with DAPI, and the images were captured using a confocal laser-scanning microscope (Olympus FV1000).
MTT assay
CNE-2 and SUNE-1 cells were seeded in 96-well plates at a density of 1,000 cells per well. At 1, 2, 3, 4, and 5 days, the cells were stained with 20 μl of MTT dye (0.5 mg/ml, Sigma) for 4 h, after which the medium was removed, and 100 μl of dimethyl sulfoxide (Sigma) was added. The absorbance was measured at 490 nm with a spectrophotometric plate reader.
Colony formation assay
CNE-2 and SUNE1 cells were seeded in six-well plates at a density of 500 cells per well and cultured for 7 or 12 days. Colonies were fixed with 4% paraformaldehyde solution, stained with 0.5% crystal violet, and counted under an inverted microscope.
Anchorage-independent soft-agar growth
CNE-2 and SUNE-1 cells (2.5 × 104) were suspended in 1 ml of complete medium containing 0.66% agar (Sigma) and then applied to the top of a 1% agar/complete medium layer in six-well plates. Colonies were counted under an inverted microscope after 9 or 12 days.
Xenograft tumor model
Three- to four-week-old male BALB/c nude mice were purchased from the Medical Experimental Animal Center of Guangdong Province (Guangzhou, China). All experimental animal protocols were approved by the Animal Care and Use Ethics Committee. SUNE-1 cells stably expressing shCIP2A or scrambled control shRNA were suspended in PBS, and 1 × 106 cells (100 μl) were subcutaneously injected into the dorsal flank of each mouse. Tumors were examined every 3 days, and tumor volumes were calculated. On day 28, the mice were sacrificed, and the tumors were dissected and weighted.
Statistical analysis
Data are presented as the mean ± SD, and differences between groups were analyzed using Student’s t-test or a chi-squared test. Receiver operation characteristic (ROC) curves were used to determine the optimal cutoff values for low and high CIP2A expression. The Kaplan-Meier method and log-rank test were used to estimate survival rates, and hazard ratios (HRs) were calculated using unadjusted univariate Cox regression analysis. Multivariate Cox regression analysis was used to test for independent prognostic factors. All statistical analyses were performed with SPSS 16.0 software, and P values of < 0.05 were considered statistically significant.
References
Xu ZJ, Zheng RS, Zhang SW, Zou XN, Chen WQ: Nasopharyngeal carcinoma incidence and mortality in China in 2009. Chin J Cancer. 2013, 32: 453-460. 10.5732/cjc.013.10118
Cho WC: Most common cancers in Asia-Pacific region: nasopharyngeal carcinoma. Cancer report of Asian-Pacific region 2010. 2010, 284-289. Asian Pacific Organization for Cancer Prevention
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D: Global cancer statistics. CA Cancer J Clin. 2011, 61: 69-90. 10.3322/caac.20107
Lai SZ, Li WF, Chen L, Luo W, Chen YY, Liu LZ, Sun Y, Lin AH, Liu MZ, Ma J: How does intensity-modulated radiotherapy versus conventional two-dimensional radiotherapy influence the treatment results in nasopharyngeal carcinoma patients?. Int J Radiat Oncol Biol Phys. 2011, 80: 661-668. 10.1016/j.ijrobp.2010.03.024
Chen Y, Sun Y, Liang SB, Zong JF, Li WF, Chen M, Chen L, Mao YP, Tang LL, Guo Y, Lin AH, Liu MZ, Ma J: Progress report of a randomized trial comparing long-term survival and late toxicity of concurrent chemoradiotherapy with adjuvant chemotherapy versus radiotherapy alone in patients with stage III to IVB nasopharyngeal carcinoma from endemic regions of China. Cancer. 2013, 119: 2230-2238. 10.1002/cncr.28049
Junttila MR, Puustinen P, Niemelä M, Ahola R, Arnold H, Böttzauw T, Ala-aho R, Nielsen C, Ivaska J, Taya Y, Lu SL, Lin S, Chan EK, Wang XJ, Grènman R, Kast J, Kallunki T, Sears R, Kähäri VM, Westermarck J: CIP2A inhibits PP2A in human malignancies. Cell. 2007, 130: 51-62. 10.1016/j.cell.2007.04.044
Khanna A, Böckelman C, Hemmes A, Junttila MR, Wiksten JP, Lundin M, Junnila S, Murphy DJ, Evan GI, Haglund C, Westermarck J, Ristimäki A: MYC-dependent regulation and prognostic role of CIP2A in gastric cancer. J Natl Cancer Inst. 2009, 101: 793-805. 10.1093/jnci/djp103
Niemelä M, Kauko O, Sihto H, Mpindi JP, Nicorici D, Pernilä P, Kallioniemi OP, Joensuu H, Hautaniemi S, Westermarck J: CIP2A signature reveals the MYC dependency of CIP2A-regulated phenotypes and its clinical association with breast cancer subtypes. Oncogene. 2012, 31: 4266-4278. 10.1038/onc.2011.599
Côme C, Laine A, Chanrion M, Edgren H, Mattila E, Liu X, Jonkers J, Ivaska J, Isola J, Darbon JM, Kallioniemi O, Thézenas S, Westermarck J: CIP2A is associated with human breast cancer aggressivity. Clin Cancer Res. 2008, 15: 5092-5100.
Li W, Ge Z, Liu C, Liu Z, Björkholm M, Jia J, Xu D: CIP2A is overexpressed in gastric cancer and its depletion leads to impaired clonogenicity, senescence, or differentiation of tumor cells. Clin Cancer Res. 2008, 14: 3722-3728. 10.1158/1078-0432.CCR-07-4137
Puustinen P, Rytter A, Mortensen M, Kohonen P, Moreira JM, Jäättelä M: CIP2A oncoprotein controls cell growth and autophagy through TORC1 activation. J Cell Biol. 2014, 204: 713-727. 10.1083/jcb.201304012
Dong QZ, Wang Y, Dong XJ, Li ZX, Tang ZP, Cui QZ, Wang EH: CIP2A is overexpressed in non-small cell lung cancer and correlates with poor prognosis. Ann Surg Oncol. 2011, 18: 857-865. 10.1245/s10434-010-1313-8
Wiegering A, Pfann C, Uthe FW, Otto C, Rycak L, Mäder U, Gasser M, Waaga-Gasser AM, Eilers M, Germer CT: CIP2A influences survival in colon cancer and is critical in maintaining Myc expression. PLoS One. 2013, 8: e75292- 10.1371/journal.pone.0075292
Yu G, Liu G, Dong J, Jin Y: Clinical implications of CIP2A protein expression in breast cancer. Med Oncol. 2013, 30: 524-
Böckelman C, Lassus H, Hemmes A, Leminen A, Westermarck J, Haglund C, Bützow R, Ristimäki A: Prognostic role of CIP2A expression in serous ovarian cancer. Br J Cancer. 2011, 105: 989-995. 10.1038/bjc.2011.346
Ren J, Li W, Yan L, Jiao W, Tian S, Li D, Tang Y, Gu G, Liu H, Xu Z: Expression of CIP2A in renal cell carcinomas correlates with tumour invasion, metastasis and patients’ survival. Br J Cancer. 2011, 105: 1905-1911. 10.1038/bjc.2011.492
Böckelman C, Hagström J, Mäkinen LK, Keski-Säntti H, Häyry V, Lundin J, Atula T, Ristimäki A, Haglund C: High CIP2A immunoreactivity is an independent prognostic indicator in early-stage tongue cancer. Br J Cancer. 2011, 104: 1890-1895. 10.1038/bjc.2011.167
Rantanen T, Kauttu T, Akerla J, Honkanen T, Krogerus L, Salo J, Paavonen T, Oksala N: CIP2A expression and prognostic role in patients with esophageal adenocarcinoma. Med Oncol. 2013, 30: 684-
Xue Y, Wu G, Wang X, Zou X, Zhang G, Xiao R, Yuan Y, Long D, Yang J, Wu Y, Xu H, Liu F, Liu M: CIP2A is a predictor of survival and a novel therapeutic target in bladder urothelial cell carcinoma. Med Oncol. 2013, 30: 406-
Lucas CM, Harris RJ, Giannoudis A, Copland M, Slupsky JR, Clark RE: Cancerous inhibitor of PP2A (CIP2A) at diagnosis of chronic myeloid leukemia is a critical determinant of disease progression. Blood. 2011, 117: 6660-6668. 10.1182/blood-2010-08-304477
Khanna A, Pimanda JE, Westermarck J: Cancerous inhibitor of protein phosphatase 2A, an emerging human oncoprotein and a potential cancer therapy target. Cancer Res. 2013, 73: 6548-6553. 10.1158/0008-5472.CAN-13-1994
Jansens V, Goris J, Hoof C: PP2A: the expected tumor suppressor. Curr Opin Genet Dev. 2005, 15: 34-41. 10.1016/j.gde.2004.12.004
Rangarajan A, Hong SJ, Gifford A, Weinberg RA: Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 2004, 6: 171-183. 10.1016/j.ccr.2004.07.009
Chen KF, Liu CY, Lin YC, Yu HC, Liu TH, Hou DR, Chen PJ, Cheng AL: CIP2A mediates effects of bortezomib on phospho-Akt and apoptosis in hepatocellular carcinoma cells. Oncogene. 2010, 29: 6257-6266. 10.1038/onc.2010.357
Kerosuo L, Fox H, Perälä N, Ahlqvist K, Suomalainen A, Westermarck J, Sariola H, Wartiovaara K: CIP2A increases self-renewal and is linked to Myc in neural progenitor cells. Differentiation. 2010, 80: 68-77. 10.1016/j.diff.2010.04.003
Tseng LM, Liu CY, Chang KC, Chu PY, Shiau CW, Chen KF: CIP2A is a target of bortezomib in human triple negative breast cancer cells. Breast Cancer Res. 2012, 14: R68- 10.1186/bcr3175
Kim JS, Kim EJ, Oh JS, Park IC, Hwang SG: CIP2A modulates cell cycle progression in human cancer cells by regulating the stability and activity of PLK1. Cancer Res. 2013, 73: 6667-6678. 10.1158/0008-5472.CAN-13-0888
Laine A, Sihto H, Come C, Rosenfeldt MT, Zwolinska A, Niemelä M, Khanna A, Chan EK, Kähäri VM, Kellokumpu-Lehtinen PL, Sansom OJ, Evan GI, Junttila MR, Ryan KM, Marine JC, Joensuu H, Westermarck J: Senescence sensitivity of breast cancer cells is defined by positive feedback loop between CIP2A and E2F1. Cancer Discov. 2013, 3: 182-197. 10.1158/2159-8290.CD-12-0292
Khanna A, Okkeri J, Bilgen T, Tiirikka T, Vihinen M, Visakirpi T, Westermarck J: ETS2 mediates MEK1/2-dependent overexpression of cancerous inhibitor of protein phosphatase 2A (CIP2A) in human cancer cells. PLoS One. 2011, 6: e17979- 10.1371/journal.pone.0017979
Mathiasen DP, Egebjerg C, Andersen SH, Rafn B, Puustinen P, Khanna A, Daugaard M, Valo E, Tuomela S, Bottzauw T, Nielsen CF, Willumsen BM, Hautaniemi S, Lahesmaa R, Westermarck J, Jäättelä M, Kallunki T: Identification of a c-Jun N-terminal kinase-2-dependent signal amplification cascade that regulates c-Myc levels in ras transformation. Oncogene. 2012, 31: 390-401. 10.1038/onc.2011.230
Pommier Y, Marchand C: Interfacial inhibitors: targeting macromolicular complexes. Nat Rev Drug Discov. 2012, 11: 25-36.
Chan AT, Teo PM, Ngan PK, Leung TW, Lau WH, Zee B, Leung SF, Cheung FY, Yeo W, Yiu HH, Yu KH, Chiu KW, Chan DT, Mok T, Yuen KT, Mo F, Lai M, Kwan WH, Choi P, Johnson PJ: Concurrent chemotherapy-dadiotherapy compared with radiotherapy alone in locoregionally advanced nasopharyngeal carcinoma: progression-free survival analysis of a phase III randomized trial. J Clin Oncol. 2002, 20: 2038-2044. 10.1200/JCO.2002.08.149
Livak KJ, Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001, 25: 402-408. 10.1006/meth.2001.1262
Liu N, Cui RX, He QM, Huang BJ, Sun Y, Xie D, Zeng J, Wang HY, Ma J: Reduced expression of Dicer1 is associated with poor prognosis in patients with nasopharyngeal carcinoma. Med Oncol. 2013, 30: 360-
Liu N, Tang LL, Sun Y, Cui RX, Wang HY, Huang BJ, He QM, Jiang W, Ma J: MiR-29c suppresses invasion and metastasis by targeting TIAM1 in nasopharyngeal carcinoma. Cancer Lett. 2013, 329: 181-188. 10.1016/j.canlet.2012.10.032
Acknowledgments
This work was supported by grants from the Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2010), the One Hundred Talents Project of Sun Yat-sen University (2013), the Innovation Team Development Plan of the Ministry of Education (No. IRT1297), the National Natural Science Foundation of China (No. 81230056), the Science and Technology Project of Guangzhou City (No. 12BppZXaa2060002), and the Key Laboratory Construction Project of Guangzhou City, China (No. 121800085).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
NL, QH, JC, YL, YX, and XR performed experiments; JM, NL, QH, JC, QL YX, XR, YS, HM, JS, WJ, TK, and MZ designed research, analyzed data and edited the manuscript for intellectual content. All authors have made critical edits to the manuscript and have given final approval.
Na Liu, Qing-Mei He, Jie-Wei Chen contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Liu, N., He, QM., Chen, JW. et al. Overexpression of CIP2A is an independent prognostic indicator in nasopharyngeal carcinoma and its depletion suppresses cell proliferation and tumor growth. Mol Cancer 13, 111 (2014). https://doi.org/10.1186/1476-4598-13-111
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1476-4598-13-111