Abstract
Background
Blood pressure (BP) is a complex trait, with a heritability of 30 to 40%. Several genome wide associated BP loci explain only a small fraction of the phenotypic variation. Family studies can provide an important tool for gene discovery by utilizing trait and genetic transmission information among relative-pairs. We have previously described a quantitative trait locus at chromosome 17q25.3 influencing systolic BP in American Indians of the Strong Heart Family Study (SHFS). This locus has been reported to associate with variation in BP traits in family studies of Europeans, African Americans and Hispanics.
Methods
To follow-up persuasive linkage findings at this locus, we performed comprehensive genotyping in the 1-LOD unit support interval region surrounding this QTL using a multi-step strategy. We first genotyped 1,334 single nucleotide polymorphisms (SNPs) in 928 individuals from families that showed evidence of linkage for BP. We then genotyped a second panel of 306 SNPs in all SHFS participants (N = 3,807) for genes that displayed the strongest evidence of association in the region, and, in a third step, included additional genotyping to better cover the genes of interest and to interrogate plausible candidate genes in the region.
Results
Three genes had multiple SNPs marginally associated with systolic BP (TBC1D16, HRNBP3 and AZI1). In BQTN analysis, used to estimate the posterior probability that any variant in each gene had an effect on the phenotype, AZI1 showed the most prominent findings (posterior probability of 0.66). Importantly, upon correction for multiple testing, none of our study findings could be distinguished from chance.
Conclusion
Our findings demonstrate the difficulty of follow-up studies of linkage studies for complex traits, particularly in the context of low powered studies and rare variants underlying linkage peaks.
Similar content being viewed by others
Background
Blood pressure (BP) is a complex trait and genetic factors account for 30 to 40% of the blood pressure variation in a population [1]. Recent progress has been made in the identification of common variants associated with BP and hypertension risk in populations [2–7], with over 50 loci for BP traits identified in genome-wide association studies (GWAS) [2–5, 8–10]. However, these findings only explain a small fraction of the phenotypic variation attributable to genetic effects [4]. Evidence for a role of rare variants in BP is well documented in monogenic forms of hypertensive and hypotensive syndromes [11, 12].
Family studies can provide an important tool for gene/loci discovery by utilizing trait and genetic transmission information among relative-pairs. Several genome scans of BP and hypertension have been published but few overlapping regions have been identified [recently reviewed in [1, 13]]. Many of these studies failed to show genome-wide significant linkage. Those demonstrating strong evidence for linkage have identified quantitative trait loci (QTLs) on chromosomes 2p [Mexican Americans [14] and families from Sardinia [15]] ,2q [African Americans [16] and Amish [17]], 3 [Finnish families [18]], 4p [Dutch families [19]], 6q [European Americans [20, 21], white Europeans [22]], 7 [African Americans [23]], 17q [European Americans [24] and Hispanics [25, 23]], 18q [Icelandic families [26] and European Americans [27]], 20 [Hispanics [23]] and 21 [European Americans [23]].
We have previously described a QTL-specific genotype-by-sex interaction for systolic BP on chromosome 17q25.3 in American Indians participants of the Strong Heart Family Study (SHFS) [28]. This is the same region identified in European Americans [24] and Hispanics [23, 25] for BP traits. This QTL became more significant when we accounted for an interaction by sex (LOD =3.4 in women in comparison to men). To follow-up persuasive linkage findings at this locus, we performed comprehensive genotyping in the chromosome 17q region using a three-stage strategy. We first genotyped a panel of single nucleotide variants (SNVs) in individuals belonging to families that showed strong evidence of linkage for systolic BP and then genotyped a second panel of SNV in all SHFS participants for the genes displaying the most prominent evidence for association in the region. As a third stage we typed additional variants in the genes of interest as well as further characterized additional candidate genes from the region. Finally, because our linkage findings were strongest in the female only sample, we examined the associations in males and females separately. Here we report the results from these analyses.
Methods
SHFS study design, population and phenotypes
We used data from the SHFS, a large family-based genetic component of the Strong Heart Study (SHS), a population-based study of cardiovascular disease and its risk factors in American Indians 45 years or older recruited from tribes in Arizona (AZ), Oklahoma (OK) and North and South Dakota (DK). The SHFS began as a pilot study in 1998 when ~900 members of extended families of the SHS cohort were examined. Additional family members were recruited from 2001 to 2003 for a total of 3,807 individuals in 94 multigenerational families (mean family size of 40 individuals, range 5 to 110). The SHFS protocols were approved by the Indian Health Services (IHS) Institutional Review Board, by Institutional Review Boards of all Institutions, and by the Indian tribes [29, 30]. All participants gave informed consent for genetic testing. The study was conducted according to the principles expressed in the Declaration of Helsinki.
Baseline socio-demographic, medical history, lifestyle and behaviors (smoking and alcohol intake) and medications were obtained through an interview using standardized questionnaires. Physical exams collected data on height, weight, systolic and diastolic BPs. Body mass index (BMI) was estimated using height and weight (kg/m2). BP was measured using a standard protocol across the three recruiting centers [30]. Brachial seated BPs were measured three times by a trained technician using a mercury column sphygmomanometer (WA Baum Co Inc, Copiague, NY) and size-adjusted cuffs. The average of the last two of the three measures was used in the analyses. Hypertension is defined by a BP of 140/90 mm Hg or higher, or use of antihypertensive drugs [31].
Genotyping strategy, methods and quality control
The genotyping strategy is shown in Additional file 1: Figure S1.
Stage 1: SHFS panel 1
SNVs were selected within the 1-LOD unit drop support interval of the chromosome 17 QTL from 69,509,00 to 77,946,426 bp (genome build 35). We identified all polymorphic variants in HapMap CEU and JPN/HCB. We used linkage disequilibrium (LD) metrics (r2) and minor allele frequency (MAF) (SNVs with a MAF <0.001 in the HapMap data were removed) to select SNVs for genotyping. We also included 2 variants located in miRNA. A total of 1,536 SNVs were genotyped in 933 SHFS participants who are members of families showing evidence of linkage for systolic BP. Of the 1,536 SNVs on chromosome 17 that were included, 1334 SNVs were heterozygous, 18 SNVs were not polymorphic, and 184 SNVs failed genotyping. Five individuals had call rates < 95% and were removed from further analyses. Therefore, 1,334 heterozygous SNVs in 920 individuals were available for analyses.
Stage 2: SHFS panel 2
To provide evidence of replication for the genes on chromosome 17 that show the highest association, we genotyped 639 of the most significant SNVs of the Panel 1 analysis in all SHFS members (n = 3,800) and included additional SNVs in regions where we had relatively low coverage in Panel 1 (n = 30 SNVs). We also genotyped additional SNVs in these genes and in several other candidate genes in the region (23 SNVs in AZI1; 156 in HRNB3; 52 in TBC1D16; 8 in ACTG1; 5 in UTS2R; 34 in ACE; 33 in SCL39A11), for a total of 980 SNVs.
Stage 3: SHS cohort genotyping
We genotyped 91 of the most prominent SNVs in additional 3,516 SHS cohort members.
For the strategies described above, genotyping was performed using the multiplex Golden Gate genotyping technology from Illumina, based on allele-specific primer extension, according to the manufacturer’s protocol (Illumina, San Diego, CA). Briefly, genomic DNA (250 ng) was activated with biotin, hybridized to a pool of locus-specific oligos. PCR amplified using fluorescent-labeled primers and hybridized to the Sentrix Array Matrix, and then fluorescence intensities were analyzed using the Illumina BeadArray Reader and BeadStudio software. Cluster calls were checked for accuracy and genotypes were exported as text files for further use in association analysis. Additional samples were typed in replica as controls for genotyping and allele calling consistencies.
Existing genetic data in the SHFS
The SHFS has existing genotypic data on ~400 microsatellite markers [32]. MAFs were derived from pedigree founders. Mendelian inconsistencies and spurious double recombinants were detected using the SimWalk2 package [33] with the overall blanking rate for both types of errors of less than 1%. Multipoint identity-by-descent (IBD) sharing was estimated using Loki [34]. Pedigree relationships were verified using the PREST package [35]. This information was used in the implementation of the Quantitative Trait Nucleotide (QTN) analysis.
Statistical analyses
We evaluated quantitative variation in systolic BP. To account for the use of anti-hypertensive medications, we added a constant to treated measures of systolic BP (10 mm Hg). Systolic BP was log-transformed due to non-normality of the data. Models also adjusted for age, sex, age2, and BMI and stratified by study center.
Association analysis
We implemented a single marker test for each SNV. To evaluate the association of SNVs with BP traits among family members, we fitted linear mixed effects models to account for within pedigree correlations (implemented in Genome-Wide Association analyses with Family [GWAF]) [36]. Genotypes were tested for additive association using a 1-df Wald test. Analyses report beta and standard error (se) per copy number of the coded allele. Summary results of each center were combined using fixed effects meta-analysis. Significant p-value thresholds were determined using a Bonferroni correction (Stage 1: 1,334 SNPs, p < 3.7 × 10−5).
Population stratification
The SHFS does not have ancestry informative markers to adjust for population stratification. Therefore, we tested for the evidence of population stratification for each variant using the quantitative transmission disequilibrium test (QTDT) [37] and a test for stratification described by Havill et al.[38], both implemented in SOLAR. To control for spurious associations due to population stratification and admixture, genotype scores are decomposed into between-family and within-family components, and the likelihood of a model in which the association parameters of these two components are estimated is compared to the likelihood of a model in which they are constrained to be equal, as expected in the absence of population stratification.
Bayesian Quantitative Trait Nucleotide (BQTN) method
For genes showing the most significant associations with systolic blood pressure, we used the BQTN to estimate the probability that each SNV is functional [39]. The BQTN method is designed to separate potentially functional variants from neutral variants in LD with them based on a displacement in the observed phenotype values and it incorporates each variant one by one, evaluating the likelihood of a model in which the trait mean varies by genotype. SNVs having a LD r2 higher than 0.90 are treated as in one group and only one SNV in each group will be used in the analysis. Bayesian model averaging/model selection was used based on additive QTN effects, for which there are 2m possible models, where m is the number of QTNs considered (m was restricted to ≤ 15 SNVs). The approach evaluates all such models and utilizes Bayesian methods to estimate the posterior probability that each SNV is functional. It then evaluates models with all possible combinations of two variants, three variants, and so on. Each model will also have the effect size estimate and its standard error for each SNV appearing in that model, the averaged effect size and standard error for that SNV in that gene and the sum of posterior model probabilities across all models containing the SNV.
Results
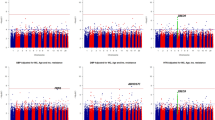
Descriptive characteristics of individuals genotyped in the SHFS and SHS are shown in Additional file 1: Table S1. The panel 1 sample was comprised of individuals from families with evidence of linkage for systolic BP (stage 1), and Panel 2 included all participants of the SHFS (Stage 2). Single SNV association results for Panel 1 are shown in Additional file 1: Table S2. Three genes had multiple SNVs marginally associated with systolic BP (TBC1D16, HRNBP3 and AZI1) and low evidence for heterogeneity across centers (Figures 1, 2 and 3). There was no evidence of population stratification for these associations (p > 0.10) except for the SNP rs8070973 in the Oklahoma sample (p = 0.03 for the stratification test). However, the p-value for association of this SNP with systolic BP was 0.008 using the QTDT test, which accounts for population stratification. The pattern of LD of these genomic regions in the three centers is shown in Additional file 1: Figure S2. These three genes were prioritized for further characterization in the second genotyping panel in the entire SHFS sample. Association findings from Panel 2 for systolic BP also pointed to the same three genes (Additional file 1: Table S3). Table 1 shows the meta-analyses main results across centers for systolic BP in these Panels and in the SHS cohort study. For SNVs genotyped in both Panels, we noticed higher effect estimates in family members selected based on the linkage results (Panel 1) compared to the overall individuals genotyped in Panel 2 (Table 2). Allele frequencies in the SHFS and SHS are shown in Additional file 1: Tables S4 and S5, respectively. Association findings for females and males are shown in Additional file 1: Tables S6 and S7.
We used the BQTN method to estimate the posterior probability that any variant in each gene had an effect on the phenotype in Panel 1 which showed stronger association estimates (Table 3). This analysis showed marginal posterior probability for a SNV on the AZI1 gene (>60%) but no strong probabilities of effects for any of the typed SNVs. BQTN analyses of Panel 2 SNVs did not reveal any SNV with a notable posterior probability of effect (data not shown).
Discussion
To follow up persuasive evidence of genome wide linkage findings for systolic BP in American Indians, we performed a comprehensive fine mapping of a chromosome 17 genomic region. We identified evidence for locus heterogeneity in association analyses, with suggestive (nominal) associations of SNVs in three genes (TBC1D16, HRNBP3 and AZI1) in single test analyses. Importantly, upon Bonferroni correction for multiple testing, none of these study findings can be distinguished from chance. Using the BQTN method to estimate the posterior probability that any SNV in each gene had an effect on the systolic BP, the AZI1 rs12939525 SNV showed the most prominent findings (posterior probability of 0.66).
The three genes that displayed the strongest evidence for association with blood pressure have not been previously associated with blood pressure traits. AZI1 encodes the 5-azacytidine induced 1 protein. This cell cycle protein is thought to play a role in spermatogenesis, through the recruitment of mitotic centrosome proteins and complexes. HRNBP3 encodes hexaribonucleotide binding protein 3. This complex gene encodes 10 different mRNAs, 9 alternatively spliced variants and 1 unspliced form. Functionally, the gene has been proposed to participate in mRNA processing/splicing and functions to bind RNA and to localize in the extracellular space, cytoplasm, and nucleus [40–42]. TBC1D16 encodes the protein TBC1 Domain Family, Member 16, which is up-regulated in melanoma. Recent studies have shown that TBC1D16 enhances the intrinsic rate of GTP hydrolysis by Rab4A, a master regulator of receptor recycling from endocytic compartments to the plasma membrane [43].
While a plausible story for a gene-phenotype relationship is often easy to make, especially given the general cell cycle functions of the identified candidate circulating proteins implicated here, no single gene nor SNV displayed a Bonferroni corrected p-value supporting statistical significance, suggesting that perhaps multiple variants of small individual effect account for the linkage evidence to chromosome 17q. Unfortunately, these findings are rather standard when viewed in the context of the many previous fine mapping studies of complex traits like blood pressure, where there has been an inherent failure to identify a single variant that accounts for a linkage peak [44, 45]. Reasons for such failures include locus heterogeneity as hypothesized here, lack of statistical power, population stratification, as well as a lack of consideration of rare variants, in particular variants with MAF < 0.01.
The sample size in these families precludes us from definitively identifying the exact set of causal genes and variants, particularly as they likely are low frequency or rare, even in this population. Novel methods are needed to map rare genes in the context of low power. These findings support the hypothesis that there are likely multiple underlying genes and/or variants and that those segregating in these pedigrees may be specific to families and not easily identified in studies of unrelated individuals. Further interrogation of these regions with sequencing to detect rare variants is warranted.
We observed differences in the patterns of SNV -BP association by sex-strata but the findings were difficult to differentiate from chance (Additional file 1: Tables S6 and S7). This is relevant because our initial linkage peak displayed evidence of sex-specific effects [28].
Conclusions
Our results illustrate the challenges of gene discovery using association analyses in the presence of locus and allelic heterogeneity, which may have implications for the study of complex traits across ancestries. American Indian population-specific variants and low frequency/rare variants not included in the HapMap were not evaluated in this study, and could account for some of the non-significant findings. The 1000 genome project data was not available at the time of SNV selection; however the American Indian gene pool is not captured very well by the 1000 genome data. Sequencing of this region could provide further information on functional SNVs at this locus. Our results also suggest that a single genetic variant is not likely to be the cause of the linkage signal on 17q for blood pressure, which represents a major challenge for variant discovery with association analysis as power to detect effects becomes much diminished.
References
Hopkins PN, Hunt SC: Genetics of hypertension. Genet Med. 2003, 5 (6): 413-429. 10.1097/01.GIM.0000096375.88710.A6.
Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, Papadakis K, Voight BF, Scott LJ, Zhang F, Farrall M, Tanaka T, Wallace C, Chambers JC, Khaw KT, Nilsson P, van der Harst P, Polidoro S, Grobbee DE, Onland-Moret NC, Bots ML, Wain LV, Elliott KS, Teumer A, Luan J, Lucas G, et al: Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009, 41 (6): 666-676. 10.1038/ng.361.
Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, Aulchenko Y, Lumley T, Kottgen A, Vasan RS, Rivadeneira F, Eiriksdottir G, Guo X, Arking DE, Mitchell GF, Mattace-Raso FU, Smith AV, Taylor K, Scharpf RB, Hwang SJ, Sijbrands EJ, Bis J, Harris TB, Ganesh SK, O'Donnell CJ, Hofman A, et al: Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009, 41 (6): 677-687. 10.1038/ng.384.
Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, Pihur V, Vollenweider P, O'Reilly PF, Amin N, Bragg-Gresham JL, Teumer A, Glazer NL, Launer L, Zhao JH, Aulchenko Y, Heath S, Sõber S, Parsa A, Luan J, Arora P, Dehghan A, Zhang F, Lucas G, Hicks AA, Jackson AU, et al: Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011, 478: 103-109. 10.1038/nature10405.
Kato N, Takeuchi F, Tabara Y, Kelly TN, Go MJ, Sim X, Tay WT, Chen CH, Zhang Y, Yamamoto K, Katsuya T, Yokota M, Kim YJ, Ong RT, Nabika T, Gu D, Chang LC, Kokubo Y, Huang W, Ohnaka K, Yamori Y, Nakashima E, Jaquish CE, Lee JY, Seielstad M, Isono M, Hixson JE, Chen YT, Miki T, Zhou X, et al: Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet. 2011, 43 (6): 531-538. 10.1038/ng.834.
Fox ER, Young JH, Li Y, Dreisbach AW, Keating BJ, Musani SK, Liu K, Morrison AC, Ganesh S, Kutlar A, Ramachandran VS, Polak JF, Fabsitz RR, Dries DL, Farlow DN, Redline S, Adeyemo A, Hirschorn JN, Sun YV, Wyatt SB, Penman AD, Palmas W, Rotter JI, Townsend RR, Doumatey AP, Tayo BO, Mosley TH, Lyon HN, Kang SJ, Rotimi CN, et al: Association of genetic variation with systolic and diastolic blood pressure among African Americans: the Candidate Gene Association Resource study. Hum Mol Genet. 2011, 20 (11): 2273-2284. 10.1093/hmg/ddr092.
Franceschini N, Reiner AP, Heiss G: Recent findings in the genetics of blood pressure and hypertension traits. Am J Hypertens. 2011, 24 (4): 392-400. 10.1038/ajh.2010.218.
Ganesh SK, Tragante V, Guo W, Guo Y, Lanktree MB, Smith EN, Johnson T, Castillo BA, Barnard J, Baumert J, Chang YP, Elbers CC, Farrall M, Fischer ME, Franceschini N, Gaunt TR, Gho JM, Gieger C, Gong Y, Isaacs A, Kleber ME, Mateo Leach I, McDonough CW, Meijs MF, Mellander O, Molony CM, Nolte IM, Padmanabhan S, Price TS, Rajagopalan R, et al: Loci influencing blood pressure identified using a cardiovascular gene-centric array. Hum Mol Genet. 2013, 22 (8): 1663-1678. 10.1093/hmg/dds555.
Wain LV, Verwoert GC, O'Reilly PF, Shi G, Johnson T, Johnson AD, Bochud M, Rice KM, Henneman P, Smith AV, Ehret GB, Amin N, Larson MG, Mooser V, Hadley D, Dorr M, Bis JC, Aspelund T, Esko T, Janssens AC, Zhao JH, Heath S, Laan M, Fu J, Pistis G, Luan J, Arora P, Lucas G, Pirastu N, Pichler I, et al: Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet. 2011, 43 (10): 1005-1011. 10.1038/ng.922.
Johnson T, Gaunt TR, Newhouse SJ, Padmanabhan S, Tomaszewski M, Kumari M, Morris RW, Tzoulaki I, O'Brien ET, Poulter NR, Sever P, Shields DC, Thom S, Wannamethee SG, Whincup PH, Brown MJ, Connell JM, Dobson RJ, Howard PJ, Mein CA, Onipinla A, Shaw-Hawkins S, Zhang Y, Davey Smith G, Day IN, Lawlor DA, Goodall AH, Fowkes FG, Abecasis GR, Elliott P, et al: Blood pressure loci identified with a gene-centric array. Am J Hum Genet. 2011, 89 (6): 688-700. 10.1016/j.ajhg.2011.10.013.
Lifton RP, Gharavi AG, Geller DS: Molecular mechanisms of human hypertension. Cell. 2001, 104 (4): 545-556. 10.1016/S0092-8674(01)00241-0.
Ji W, Foo JN, O'Roak BJ, Zhao H, Larson MG, Simon DB, Newton-Cheh C, State MW, Levy D, Lifton RP: Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008, 40 (5): 592-599. 10.1038/ng.118.
Samani NJ: Genome scans for hypertension and blood pressure regulation. Am J Hypertens. 2003, 16 (2): 167-171. 10.1016/S0895-7061(02)03244-2.
Atwood LD, Samollow PB, Hixson JE, Stern MP, MacCluer JW: Genome-wide linkage analysis of blood pressure in Mexican Americans. Genet Epidemiol. 2001, 20 (3): 373-382. 10.1002/gepi.7.
Angius A, Petretto E, Maestrale GB, Forabosco P, Casu G, Piras D, Fanciulli M, Falchi M, Melis PM, Palermo M, Pirastu M: A new essential hypertension susceptibility locus on chromosome 2p24-p25, detected by genomewide search. Am J Hum Genet. 2002, 71 (4): 893-905. 10.1086/342929.
Morrison AC, Cooper R, Hunt S, Lewis CE, Luke A, Mosley TH, Boerwinkle E: Genome scan for hypertension in nonobese African Americans: the National Heart, Lung, and Blood Institute Family Blood Pressure Program. Am J Hypertens. 2004, 17 (9): 834-838.
Hsueh WC, Mitchell BD, Schneider JL, Wagner MJ, Bell CJ, Nanthakumar E, Shuldiner AR: QTL influencing blood pressure maps to the region of PPH1 on chromosome 2q31-34 in Old Order Amish. Circulation. 2000, 101 (24): 2810-2816. 10.1161/01.CIR.101.24.2810.
Perola M, Kainulainen K, Pajukanta P, Terwilliger JD, Hiekkalinna T, Ellonen P, Kaprio J, Koskenvuo M, Kontula K, Peltonen L: Genome-wide scan of predisposing loci for increased diastolic blood pressure in Finnish siblings. J Hypertens. 2000, 18 (11): 1579-1585. 10.1097/00004872-200018110-00008.
Allayee H, de Bruin TW, Michelle Dominguez K, Cheng LS, Ipp E, Cantor RM, Krass KL, Keulen ET, Aouizerat BE, Lusis AJ, Rotter JI: Genome scan for blood pressure in Dutch dyslipidemic families reveals linkage to a locus on chromosome 4p. Hypertension. 2001, 38 (4): 773-778. 10.1161/hy1001.092617.
Krushkal J, Ferrell R, Mockrin SC, Turner ST, Sing CF, Boerwinkle E: Genome-wide linkage analyses of systolic blood pressure using highly discordant siblings. Circulation. 1999, 99 (11): 1407-1410. 10.1161/01.CIR.99.11.1407.
Hunt SC, Ellison RC, Atwood LD, Pankow JS, Province MA, Leppert MF: Genome scans for blood pressure and hypertension: the National Heart, Lung, and Blood Institute Family Heart Study. Hypertension. 2002, 40 (1): 1-6. 10.1161/01.HYP.0000022660.28915.B1.
Caulfield M, Munroe P, Pembroke J, Samani N, Dominiczak A, Brown M, Benjamin N, Webster J, Ratcliffe P, O'Shea S, Papp J, Taylor E, Dobson R, Knight J, Newhouse S, Hooper J, Lee W, Brain N, Clayton D, Lathrop GM, Farrall M, Connell J: Genome-wide mapping of human loci for essential hypertension. Lancet. 2003, 361 (9375): 2118-2123. 10.1016/S0140-6736(03)13722-1.
Bielinski SJ, Lynch AI, Miller MB, Weder A, Cooper R, Oberman A, Chen YD, Turner ST, Fornage M, Province M, Arnett DK: Genome-wide linkage analysis for loci affecting pulse pressure: the Family Blood Pressure Program. Hypertension. 2005, 46 (6): 1286-1293. 10.1161/01.HYP.0000191706.41980.29.
Levy D, DeStefano AL, Larson MG, O'Donnell CJ, Lifton RP, Gavras H, Cupples LA, Myers RH: Evidence for a gene influencing blood pressure on chromosome 17. Genome scan linkage results for longitudinal blood pressure phenotypes in subjects from the framingham heart study. Hypertension. 2000, 36 (4): 477-483. 10.1161/01.HYP.36.4.477.
Kraja AT, Rao DC, Weder AB, Cooper R, Curb JD, Hanis CL, Turner ST, Andrade MD, Hsiung CA, Quertermous T, Zhu X, Province MA: Two Major QTLs and Several Others Relate to Factors of Metabolic Syndrome in the Family Blood Pressure Program. Hypertension. 2005, 46 (4): 751-757. 10.1161/01.HYP.0000184249.20016.bb.
Kristjansson K, Manolescu A, Kristinsson A, Hardarson T, Knudsen H, Ingason S, Thorleifsson G, Frigge ML, Kong A, Gulcher JR, Stefansson K: Linkage of essential hypertension to chromosome 18q. Hypertension. 2002, 39 (6): 1044-1049. 10.1161/01.HYP.0000018580.24644.18.
Pankow JS, Rose KM, Oberman A, Hunt SC, Atwood LD, Djousse L, Province MA, Rao DC: Possible locus on chromosome 18q influencing postural systolic blood pressure changes. Hypertension. 2000, 36 (4): 471-476. 10.1161/01.HYP.36.4.471.
Franceschini N, MacCluer JW, Goring HH, Cole SA, Rose KM, Almasy L, Diego V, Laston S, Lee ET, Howard BV, Best LG, Fabsitz RR, Roman MJ, North KE: A quantitative trait loci-specific gene-by-sex interaction on systolic blood pressure among American Indians: the Strong Heart Family Study. Hypertension. 2006, 48 (2): 266-270. 10.1161/01.HYP.0000231651.91523.7e.
North KE, Williams JT, Welty TK, Best LG, Lee ET, Fabsitz RR, Howard BV, MacCluer JW: Evidence for joint action of genes on diabetes status and CVD risk factors in American Indians: the strong heart family study. Int J Obes Relat Metab Disord. 2003, 27 (4): 491-497. 10.1038/sj.ijo.0802261.
Lee ET, Welty TK, Fabsitz R, Cowan LD, Le NA, Oopik AJ, Cucchiara AJ, Savage PJ, Howard BV: The Strong Heart Study. A study of cardiovascular disease in American Indians: design and methods. Am J Epidemiol. 1990, 132 (6): 1141-1155.
Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jones DW, Materson BJ, Oparil S, Wright JT, Roccella EJ: The Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure: the JNC 7 report. Jama. 2003, 289 (19): 2560-2572. 10.1001/jama.289.19.2560.
North KE, Almasy L, Goring HH, Cole SA, Diego VP, Laston S, Cantu T, Williams JT, Howard BV, Lee ET, Best LG, Fabsitz RR, MacCluer JW: Linkage analysis of factors underlying insulin resistance: Strong Heart Family Study. Obes Res. 2005, 13 (11): 1877-1884. 10.1038/oby.2005.230.
Sobel E, Papp JC, Lange K: Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet. 2002, 70 (2): 496-508. 10.1086/338920.
Heath SC, Snow GL, Thompson EA, Tseng C, Wijsman EM: MCMC segregation and linkage analysis. Genet Epidemiol. 1997, 14 (6): 1011-1016. 10.1002/(SICI)1098-2272(1997)14:6<1011::AID-GEPI75>3.0.CO;2-L.
Sun L, Wilder K, McPeek MS: Enhanced pedigree error detection. Hum Hered. 2002, 54 (2): 99-110. 10.1159/000067666.
Chen MH, Yang Q: GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010, 26 (4): 580-581. 10.1093/bioinformatics/btp710.
Abecasis GR, Cardon LR, Cookson WO: A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000, 66 (1): 279-292. 10.1086/302698.
Havill LM, Dyer TD, Richardson DK, Mahaney MC, Blangero J: The quantitative trait linkage disequilibrium test: a more powerful alternative to the quantitative transmission disequilibrium test for use in the absence of population stratification. BMC Genet. 2005, 6 (Suppl 1): S91-10.1186/1471-2156-6-S1-S91.
Blangero J, Goring HH, Kent JW, Williams JT, Peterson CP, Almasy L, Dyer TD: Quantitative trait nucleotide analysis using Bayesian model selection. Hum Biol. 2005, 77 (5): 541-559. 10.1353/hub.2006.0003.
Kim KK, Adelstein RS, Kawamoto S: Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J Biol Chem. 2009, 284 (45): 31052-31061. 10.1074/jbc.M109.052969.
Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL: Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005, 25 (22): 10005-10016. 10.1128/MCB.25.22.10005-10016.2005.
Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, et al: The sequence of the human genome. Science. 2001, 291 (5507): 1304-1351. 10.1126/science.1058040.
Goueli BS, Powell MB, Finger EC, Pfeffer SR: TBC1D16 is a Rab4A GTPase activating protein that regulates receptor recycling and EGF receptor signaling. Proc Natl Acad Sci U S A. 2012, 109 (39): 15787-15792. 10.1073/pnas.1204540109.
Ehret GB, O'Connor AA, Weder A, Cooper RS, Chakravarti A: Follow-up of a major linkage peak on chromosome 1 reveals suggestive QTLs associated with essential hypertension: GenNet study. Eur J Hum Genet. 2009, 17 (12): 1650-1657. 10.1038/ejhg.2009.94.
Das SK, Chu W, Zhang Z, Hasstedt SJ, Elbein SC: Calsquestrin 1 (CASQ1) gene polymorphisms under chromosome 1q21 linkage peak are associated with type 2 diabetes in Northern European Caucasians. Diabetes. 2004, 53 (12): 3300-3306. 10.2337/diabetes.53.12.3300.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2261/14/158/prepub
Acknowledgments
R01HL089651; The Strong Heart Study and Strong Heart Family Study were funded though the following NIH grants: R01HL041654, R01HL041642, R01HL041652, R01HL065520, and R01HL065521. This investigation was partially conducted in facilities constructed with support for the NIH Research Facilities Improvement Program Grant Number C06RR013556.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
NF, SAC and KEN designed the study and wrote the manuscript. RT, NF and LL performed the statistical analysis. KH, SAC, SR and SL genotyped the SNPs. ETL, LA, LGB, RF contributed generating the phenotype data. SR, KH, LA, HHHG, SL, ETL, LGB, RF contributed with critical review of the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
12872_2014_802_MOESM1_ESM.pdf
Additional file 1: Genotyping strategy for fine mapping of the chromosome 17 locus using family (SHFS) and cohort (SHS) data.(PDF 4 MB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Franceschini, N., Tao, R., Liu, L. et al. Mapping of a blood pressure QTL on chromosome 17 in American Indians of the strong heart family study. BMC Cardiovasc Disord 14, 158 (2014). https://doi.org/10.1186/1471-2261-14-158
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2261-14-158