Abstract
Background
During June-July 2012, six imipenem-resistant Escherichia coli isolates were isolated from two patients hospitalized in a ward of one large tertiary-care hospital in Genoa, Italy. Genetic features associated with bla NDM-4 gene were investigated.
Results
The isolates exhibited the same PFGE profile and a multidrug-resistant (MDR) phenotype to aminoglycosides, fluoroquinolones, and β-lactams. The strains produced the NDM-4 carbapenemase and the bla NDM-4 gene was part of the variable region of a class 1 integron. MLST analysis revealed that all isolates belonged to sequence type 405 (ST405).
Conclusions
This is the first report on the emergence of an MDR strain of E.coli producing the NDM-4 MBL in Italy.
Similar content being viewed by others
Background
The emerging New Delhi metallo-β-lactamase (NDM), an acquired class B carbapenemase that was first detected in Klebsiella pneumoniae isolate from a Swedish patient of Indian origin has become a major public health concern worldwide [1].
Two cases of the new variant, NDM-4, have been recently described in isolated recovered from patients previously hospitalized in India and Cameroon [2, 3]. In Italy, a few cases of NDM-1 producing E.coli and K.pneumoniae have been reported during 2009 and 2011 [4, 5].
In this study we described six NDM-4-producing E.coli isolates obtained from two patients admitted to an Italian hospital. We also present data on the localization and the genetic environment of the bla NDM-4 gene.
Methods
Bacterial strains
Six E.coli isolated from urine samples of two inpatients at the San Martino-IST University Hospital (Genoa, Italy) were studied. Isolates were taken as part of standard patient care and informed consent for the use of clinical data has been obtained by both patients.
Strain identification, antibiotic susceptibility testing and phenotypic screening for MBL production
Routine identification and antibiotic susceptibility testing were carried out using the Vitek-2 automated system (BioMérieux, Marcy-L’etoile, France). In vitro activity of carbapenems, aztreonam, fosfomycin and nitrofurantoin was further determined by the broth microdilution method and interpreted according to the of European Committee on Antimicrobial Susceptibility Testing (EUCAST ) guidelines (Version 4.0, 2014) [6]. To detect metallo-β-lactamase (MBL) production, a synergy test using imipenem and EDTA discs was used [7].
Pulsed-field gel electrophoresis (PFGE)
Genomic DNA was prepared, digested with XbaI (New England Biolabs Inc., MA, USA) and subjected to PFGE with the CHEF DRII device (Bio-rad, Milan, Italy), as described previously [8]. Fingerprinting pattern was interpreted according to the method of Tenover et al. [9].
Multilocus sequence typing (MLST)
MLST was carried out using protocols and conditions described on the E.coli MLST website (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli/documents/primersColi_html). Sequence types were assigned using the website interface.
Molecular analysis techniques
Polymerase chain reaction (PCR) amplification of the bla NDM gene and direct sequencing of the PCR products was performed as previously described [10]. Screening for resistance genes was carried out using primers and conditions previously described [11–13]. Phylogenetic analysis using multiplex PCR method as described previously [14] was used. PCR experiments were performed to identify the upstream- and downstream-located regions of the bla NDM-4 gene [15]. Mapping of the variable region of class 1 integron was performed by PCR as described previously [16]. The genetic environment of bla NDM-4 was studied by PCR mapping and sequencing as described previously [13].
Conjugation assay and plasmid study
Plasmid transfer was attempted by conjugation, using E.coli J53 as the recipient, as described previously [17]. Plasmid DNA, isolated from E.coli, was obtained by the alkaline lysis method and was used as a template in PCR analysis with primers that are specific for bla NDM and bla CTX-M[18]. To rule out chromosomal DNA contamination the template was used to amplify an internal fragment of the house-keeping recA gene. A PCR-based replicon typing method was used to identify the incompatibility group [19].
Results
Bacteria and patients
The first NDM-4-positive E.coli isolate (URO734, index strain) was detected from the urine of a 61-year-old male inpatient (patient 1) of the rehabilitation unit of the San Martino-IST Hospital on 30 June 2012 (Figure 1). At the beginning of June, the patient was hospitalized for 7 days, in a hospital in New Delhi, India, with a history of right middle cerebral artery ischemic stroke and left-sided hemiparesis. On 15 June 2012 the patient was admitted to San Martino-IST stroke center and on 26 June he was transferred in the rehabilitation unit for 57 days. Subsequent urine samples, collected during the hospitalization period (9 July, 12 July, 27 July), continued to yield NDM-4-positive E. coli showing the same MDR phenotype as URO734 until 27 July. The patient was empirically treated with colistin. Subsequent urine samples (03 August, 09 August) were negative for E. coli.
A second case of urinary tract infection sustained by NDM-4-positive E. coli was detected in July 2012 in another inpatient (patient 2), a 79-year-old man, with a history of hip replacement, who was admitted to the same rehabilitation unit during a period overlapping the admittance of the index case. The first isolate from patient 2 (isolate URO735) was contemporary with the second isolate from patient 1. Subsequent urine sample, collected during the admission period (17 July), continued to yield NDM-4-positive E. coli, showing the same MDR phenotype as URO734. Initially, the patient was empirically treated with pipemidic acid and then, after antimicrobial susceptibility results were available, with nitrofurantoin. The clinical condition of the patient improved and the patient was discharged, without further positive urine culture. No history of travel in India or other NDM endemic areas was reported for this patient.
Antimicrobial susceptibility
The NDM-4-positive E. coli isolates exhibited a MDR phenotype to aminoglycosides, fluoroquinolones, and all β-lactams tested. The strains were susceptible to colistin, nitrofurantoin, fosfomycin and tigecycline (Table 1). All NDM-4-positive isolates produced metallo-β-lactamase (MBL) activity by the imipenem-EDTA double-disk synergy test.
Phylogenetic group and PFGE
E. coli can be classified as phylogroup A, B1, B2 or D according to the phylogenetic relationship of the sequences. Phylogenetic analysis showed that isolates belonged to the phylogenetic group D, which includes extra-intestinal isolate. All isolates exhibited the same PFGE macrorestriction profile (Figure 2).
MLST
All the NDM4-positive isolates were designated to a certain MLST sequence type by the combination of the seven allelic housekeeping genes. MLST analysis revealed that all isolates belonged to sequence type 405 (ST405).
Genetic context of bla NDM4
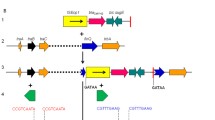
In the index isolate, PCR and sequencing analysis detected the presence of bla NDM-4 and of the following acquired resistance genes: bla TEM-1, bla CTX-M-15, dfrA12, aac (3)-II, aadA2. No other carbapenemase genes (OXA-48 or VIM types) were identified in these isolate. The resistance determinants dfrA12 and aadA2 were carried on gene cassette inserted into a class 1 integron (Figure 3), resulting in a cassette array identical to that previously described in E.coli GUE-NDM isolate from India (accession number JQ364967).
Genetic structures surrounding the bla NDM-4 gene performed by PCR identified immediately upstream of the gene the ISAba125 insertion sequence and downstream of the gene was identified the ble MBL gene encoding the resistance to bleomycin (Figure 3).
Plasmid features
The bla NDM gene could not be transferred by conjugation to E.coli J53 recipient. All strains carried a large plasmid (>23 Kb) and when the plasmid band was extracted from the gel and used as templates for the amplification of the bla NDM and bla CTX-M genes, the specific products were detected, suggesting that both resistance determinants resided in this plasmid. The PCR-based replicon typing method showed that bla NDM-4 -positive plasmid belonged to the IncF incompatibility group.
Discussion
In this communication, we described the first isolation of NDM-4 producing E.coli in Italy, represented by E.coli of sequence type 405(ST405). E.coli ST405 belonging to phylogenetic group D is increasingly reported as multidrug resistant strains causing extra-intestinal infections [20] and is a well-known pandemic clonal lineage implicated as vehicles driving the international spread of bla CTX-M [21].
NDM is not associated with certain clones, plasmids or transposons [13], our bla NDM-4 -positive plasmid belonged to the IncF incompatibility group which is known to be a major vehicle for dissemination of the bla CTX-M-15 gene [22].
By analyzing genetic structures surrounding the bla NDM-4 gene, we identified insertion sequence ISAba125 upstream and the bleomycin resistance gene downstream of the bla NDM-4 gene, this genetic environment of our NDM isolates was the same observed for most NDM-1 positive enterobacterial isolates [13].
In both patients, after treatment with in vitro active antimicrobial agents (colistin and nitrofurantoin), clinical improvement was observed and in subsequent urine samples of patient 1 E. coli NDM-4 was no longer isolated. Patient 2 was discharged without further microbiological investigation.
Patient 1 was previously hospitalized in India, a geographical region with high prevalence of NDM-producing isolates. This is the first example of importation of an Indian NDM-4-producing isolate in Italy following a hospital transfer, confirming the recent observations suggesting that the Indian subcontinent may represent an important reservoir of NDM producers. Because patient 2 had not a history of travel to NDM endemical areas and PFGE profile of the strains was identical, it is plausible that a spread of NDM-4 -producing E.coli from patient 1 to patient 2 occurred.
According to the hospital microbiology laboratory records, no further isolation of NDM-4-positive bacteria was reported to date in our hospital. To our knowledge, we report here the first NDM-4 producing E.coli detected in Italy and the fourth worldwide [2, 3, 23] . NDM-4 producing E.coli strains have been previously described in patients from India, Cameroon and Denmark. In this last case, the Danish patient was previously hospitalizes in Vietnam. In three cases (Cameroon, Denmark and Italy), isolates belonged to the ST405 sequence type. This finding is alarming because, ST405, has been previously identified as a successful international sequence type and it could favor the spread of NDM producers.
Conclusions
This is the first report on the emergence of an MDR strain of E.coli producing the NDM-4 MBL in Italy as the result of importation of an Indian NDM-4-producing isolate following a hospital transfer. The isolate belonged to a well-known international sequence type (ST405) able to spread and cause outbreak.
Our data confirms the need for a systematic screening to rapidly detect NDM-producing strains especially among patients previously hospitalized in the endemic geographic areas to avoid dissemination of carbapenemase-producing Enterobacteriaceae.
Authors’ information
Erika Coppo is a Microbiology PhD student working at the Microbiology Unit, DISC, University of Genoa, Italy. Valerio Del Bono, MD, has been working since 1994 in Infectious Disease department in Genoa as attending physician in chief. He is a member, as a responsible for Infectious Disease, of the healthcare-associated infection control team of San Martino-IST Hospital. He acts as a referee for several international journals. He is author or co-author of more 40 internationally published papers. Francesco Ventura is the head of the rehabilitation Unit, IRCCS AOU San Martino-IST, Genoa. Marco Camera is a specialist in Infectious Disease working at the IRCCS AOU San Martino-IST, Genoa Giovanni Orengo was medical director of San Martino-IST Hospital until 2011 and then director of hospital hygiene to date. His main goal was to implement active microbiological surveillance systems and he’s director of the Committee for the fight against Nosocomial Infections. Claudio Viscoli is Full Professor of Infectious Diseases at the University of Genoa, Genoa, Italy. He is the head of the Infectious Diseases Unit, IRCCS AOU San Martino-IST, Genoa. He had published more than 100 international papers. Anna Marchese is Associate Professor of Clinical Microbiology at the University of Genoa, Genoa, Italy. Her research fields include: epidemiology of mechanisms of antibiotic resistance, antimicrobial susceptibility testing, antimicrobial profile of new drugs, bacterial genetics. She has published more than 80 international papers.
Abbreviations
- NDM:
-
New Dehli Metallo-beta-Lactamase
- MBL:
-
Metallo-beta-Lactamase
- PFGE:
-
Pulse-Field Gel Electrophoresis
- PCR:
-
Polymerase Chain Reaction
- MDR:
-
Multidrug resistant
- EUCAST:
-
European Committee on Antimicrobial Susceptibility Testing
- MLST:
-
Multi Locus Sequence Typing
- MIC:
-
Minimum Inhibitory Concentration.
References
Bonomo RA: New Delhi metallo-beta-lactamase and multidrug resistance: a global SOS?. Clin Infect Dis. 2011, 52: 485-487. 10.1093/cid/ciq179.
Nordmann P, Boulanger AE, Poirel L: NDM-4 metallo-β-lactamase with increased carbapenemase activity from Escherichia coli. Antimicrob Agents Chemother. 2012, 56: 2184-2186. 10.1128/AAC.05961-11.
Dortet L, Poirel L, Anguel N, Nordmann P: New Dehli metallo- β-lactamase 4-producing Escherichia coli in Cameroon. Emerg Infect Dis. 2012, 18: 1540-1542. 10.3201/eid1809.120011.
D’Andrea MM, Venturelli C, Giani T, Arena F, Conte V, Bresciani P, Rumpianesi F, Pantosti A, Narni F, Rossolini GM: Persistent carriage and infection by multidrug-resistant Escherichia coli ST405 producing NDM-1 carbapenemase: report on the first italian cases. J Clin Microbiol. 2011, 49 (Suppl 7): 2755-2758.
Gaibani P, Ambretti S, Berlingeri A, Cordovana M, Farruggia P, Panico M, Landini MP, Sambri V: Outbreak of NDM-1-producing Enterobacteriaceae in northen Italy, July to August 2011. Euro Surveill. 2011, 16 (Suppl 47): 20027-
The European Committee on Antimicrobial susceptibility testing: Breakpoint tables for interpretation of MIC’s and zone diameters. 2014, Version 4.0, 2014. http://www.eucast.org
Arakawa Y, Shibata N, Shibayama K, Kurokawa H, Yagi T, Fujiwara H, Goto M: Convenient test for screening metallo-β-lactamase-producing gram-negative bacteria by using thiol compounds. J Clin Microbiol. 2000, 38: 40-43.
Yuan M, Aucken H, Hall LM, Pitt TL, Livermore DM: Epidemiological typing of Klebsiella with extended-spectrum β-lactamases from European intensive care units. J Antimicrob Chemother. 1998, 41: 527-539. 10.1093/jac/41.5.527.
Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B: Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995, 33: 2233-2239.
Hornsey M, Phee L, Wareham DW: A novel variant, NDM-5, of New Delhi metallo beta lactamase in a multidrug resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother. 2011, 55 (Suppl 12): 5952-5954.
Pagani L, Dell’Amico E, Migliavacca R, D’Andrea MM, Giacobone E, Amicosante G, Romero E, Rossolini GM: Multiple CTX-M-type extended-spectrum beta-lactamases in nosocomial isolates of Enterobacteriaceae from a hospital in northern Italy. J Clin Microbiol. 2003, 41 (Suppl 9): 4264-4269.
Sáenz Y, Briñas L, Domínguez E, Ruiz J, Zarazaga M, Vila J, Torres C: Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob Agents Chemother. 2004, 48 (Suppl 10): 3996-4001.
Poirel L, Dortet L, Bernabeu S, Nordmann P: Genetic features of bla NDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother. 2011, 54 ((Suppl 11)): 5403-5407.
Clermont O, Bonacorsi S, Bingen E: Rapid and sample determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000, 66: 4555-4558. 10.1128/AEM.66.10.4555-4558.2000.
Poirel L, Lagrutta E, Taylor P, Pham J, Nordmann P: Emergence of metallo-β-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob Agents Chemother. 2010, 54: 4914-4916. 10.1128/AAC.00878-10.
Lévesque C, Roy P: PCR analysis of integrons, p. 590–594 In Persing. Diagnostic molecular microbiology:principles and application. Edited by: Smith DHTF, Tenover FC, White TJ. 1993, Washington, DC: American Society for Microbiology
Lauretti L, Riccio ML, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, Rossolini GM: Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother. 1999, 43 (Suppl 7): 1584-1590.
Tokatlidou D, Tsivitanidou M, Pournaras S, Ikonomidis A, Tsakris A, Sofianou D: Outbreak caused by a multidrug-resistant Klebsiella pneumoniae clone carrying bla VIM-12 in a University hospital. J Clin Microbiol. 2008, 46 (Suppl 3): 1005-1008.
Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ: Identification of plasmids by PCR based replicon typing. J Microbiol Methods. 2005, 63: 219-228. 10.1016/j.mimet.2005.03.018.
Novais A, Vuotto C, Pires J, Montenegro C, Donelli G, Coque TM, Peixe L: Diversity and biofilm-production ability among isolates of Escherichia coli phylogroup D belonging to ST69, ST393 and ST405 clonal groups. BMC Microbiol. 2013, 13: 144-10.1186/1471-2180-13-144.
Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, Peixe L, Baquero F, Cantón R, Nordmann P: Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg Infect Dis. 2008, 14 (Suppl 2): 195-200.
Boyd DA, Tyler S, Christianson S, McGeer A, Muller MP, Willey BM, Bryce E, Gardam M, Nordmann P, Mulvey MR: Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob Agents Chemother. 2004, 48 (Suppl 10): 3758-3764.
Jakobsen L, Hammerum AM, Hansen F, Fuglsang-Damgaard D: An ST405 NDM-4-producing Escherichia coli isolated from a Danish patient previously hospitalized in Vietnam. J Antimicrob Chemother. 2014, 69 (Suppl 2): 559-560.
Acknowledgements
We would like to thank O. Varnier, Head of the Diagnostic Microbiology Unit. We gratefully acknowledge P. Gritti for her technical diagnostic assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
EC carry out the experiments AM carried out microbiological diagnostic analysis, designed the study and wrote the manuscript; FV, VDB and MC produced clinical and infectious diseases data and revised the manuscript, GO implemented microbiological procedures to detect carbapenemase producing strains and monitored their emergence during the study period. CV critically revised the manuscript. All authors read and approved the final version for publication.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Coppo, E., Del Bono, V., Ventura, F. et al. Identification of a New Delhi metallo-β-lactamase-4 (NDM-4)-producing Escherichia coli in Italy. BMC Microbiol 14, 148 (2014). https://doi.org/10.1186/1471-2180-14-148
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-14-148