Abstract
Background
Chemoreception is based on the senses of smell and taste that are crucial for animals to find new food sources, shelter, and mates. The initial step in olfaction involves the translocation of odorants from the periphery through the aqueous lymph of the olfactory sensilla to the odorant receptors most likely by chemosensory proteins (CSPs) or odorant binding proteins (OBPs).
Results
To better understand the roles of CSPs and OBPs in a coleopteran pest species, the red flour beetle Tribolium castaneum (Coleoptera, Tenebrionidae), we performed transcriptome analyses of male and female antennae, heads, mouthparts, legs, and bodies, which revealed that all 20 CSPs and 49 of the 50 previously annotated OBPs are transcribed. Only six of the 20 CSP are significantly transcriptionally enriched in the main chemosensory tissues (antenna and/or mouthparts), whereas of the OBPs all eight members of the antenna binding proteins II (ABPII) subgroup, 18 of the 20 classic OBP subgroup, the C + OBP, and only five of the 21 C-OBPs show increased chemosensory tissue expression. By MALDI-TOF-TOF MS protein fingerprinting, we confirmed three CSPs, four ABPIIs, three classic OBPs, and four C-OBPs in the antennae.
Conclusions
Most of the classic OBPs and all ABPIIs are likely involved in chemoreception. A few are also present in other tissues such as odoriferous glands and testes and may be involved in release or transfer of chemical signals. The majority of the CSPs as well as the C-OBPs are not enriched in antennae or mouthparts, suggesting a more general role in the transport of hydrophobic molecules.
Similar content being viewed by others
Background
The red flour beetle Tribolium castaneum (Herbst, Coleoptera, Tenebrionidae) is a secondary pest of stored, dried food products [1]. As a coleopteran model system, it represents the largest insect order, containing many different pests like bark beetles (Dendroctonus ponderosae, Ips typographus), colorado potato beetle (Leptinotarsa decemlineata), pollen beetle (Brassicogethes aeneus) and the Western corn rootworm (Diabrotica virgifera), which cause severe economic and ecological damage. Over the past years, T. castaneum turned into a remarkable model organism with plenty of genetic tools such as systemic RNA interference [2, 3], forward genetics based on insertional mutagenesis [4], transgene-based mis-expression systems [5, 6], as well as a fully annotated genome sequence [7, 8]. These tools predestine T. castaneum as a model system for coleopterans and to investigate findings from the vinegar fly Drosophila melanogaster for their generality in insects.
Odor discrimination is a key process in insect life: from food and host finding to partner recognition, insects rely strongly on odor stimuli. Perception of odorants takes place in the chemosensory (olfactory or gustatory) sensilla and is supposed to be mediated by chemosensory proteins (CSPs) or odorant binding proteins (OBPs) [9–13], followed by detection via odorant receptors (ORs), ionotropic glutamate-like receptors (IRs), or gustatory receptors (GRs) [14]. The olfactory sensilla are hair like structures with the highest density on the antennae. They are housing the dendrites of the odorant receptor neurons and are filled with aqueous lymph. This lymph is secreted by non-neuronal auxiliary cells and contains some CSPs and OBPs [15, 16]. The CSPs and OBPs are small (10 to 30 kDa), globular, and water soluble proteins [17] providing a hydrophobic pocket for ligand binding [18]. The CSPs are characterized by four, conserved cysteine residues forming two disulfide bonds (C1-C2, C3–C4) [19]. The OBPs – classic OBPs and antennal binding proteins (ABPIIs) [17] – have six highly conserved cysteine residues forming three interlocking disulfide bonds (C1–C3, C2–C5, C4–C6) between six α-helices, conferring a high stability to these proteins [18, 20]. The C-OBPs seem to be derived from classic OBPs and are lacking the C2–C5 disulfide bridge [17, 21–23].
It is believed that hydrophobic semiochemicals interact first with CSPs or OBPs to get shuttled through the aqueous sensillar lymph and to finally reach and activate ORs [14]. Besides evidence that CSPs are involved in chemoreception of the alfalfa plant bug Adelphocoris lineolatus and the Japanese carpenter ant Camponotus japonicus[24, 25] and their presence in the antennae of various species [10, 13, 26–29], there are no functional experiments conducting a role in chemo-sensation. In contrast, the involvement of OBPs in olfaction has been verified by several functional studies: experiments conducted with moth pheromone receptors in heterologous expression systems [30–32] or in vivo using the Drosophila melanogaster “empty neuron system” [33, 34] revealed that the presence of the corresponding OBP (pheromone binding protein, PBP) increases the sensitivity to the pheromone by 2 to 3 orders of magnitude (reviewed in [14]). Additionally, D. melanogaster mutants for the OBP Lush [35], allelic variation of different OBPs in D. melanogaster[36] and of an OBP in the fire ant Solenopsis invicta[37], as well as several RNAi based experiments in D. melanogaster and mosquitoes [38–40] showed that OBPs are important for the correct and highly sensitive reception of different semiochemicals in these insects, but might not be absolutely essential [41]. However, expression analysis of different insects have revealed that CSPs and OBPs are not restricted to the main chemosensory tissues [42–48] but are also involved in other tasks, e.g. the release of semiochemicals [49], mating [50], embryogenesis [51], immune-response [52], and regeneration [53]. Moreover, olfactory based systems [54] such as OBP coupled biosensors might improve pest and plant disease monitoring [55, 56], risk assessment [57], or prevent infestation by camouflaging or repelling [58, 59]. This could offer novel eco-friendly and cost effective ways to combat the fast adaption of Tribolium against several insecticides and respective resistance development [60, 61] and thus improve the protection of stored agricultural products [62] against migrating beetles [63].
In this study we use tissue-specific transcriptomics to improve the genome annotation of the T. castaneum CSPs and OBPs and to determine their expression profile. We place these data into a phylogenetic context in order to get better insights into their potential functions with a comparative evolutionary perspective.
Methods
Triboliumrearing
T. castaneum strain San Bernardino (Herbst, 1797; Insecta, Coleoptera, Tenebrionidae), was reared on organic wheat flour supplemented with 5% yeast powder at 28°C and 40% relative humidity under constant light. The Beetles were collected from different breeding boxes varying in age (up to three month) and culture density.
RNA isolation and sequencing
From the sex separated and age pooled animals about 1000 antennae, 600 legs, 150 mouthparts (as piece of the head capsule anterior of the antennae), 50 heads (the whole head capsule excluding the antennae) and 20 bodies (excluding head and legs) were manually dissected and immediately transferred to ice cold RNA lysis buffer (Zymo Research, Irvine, USA). For larval tissues about 100 heads and 50 bodies of unsexed last instar larvae were collected. Total RNA was isolated using the ZR Tissue & Insect RNA Micro Prep Kit (Zymo Research, Freiburg, Germany) following the manufacturer’s protocol. The Library preparation for RNA-Seq was performed using the TruSeq RNA Sample Preparation Kit (Illumina, San Diego, USA) starting from 300 ng of total RNA. Accurate quantification of cDNA libraries was performed by using the QuantiFluor™ dsDNA System (Promega, Fitchburg, USA). The size range of final cDNA libraries (280 bp) was determined applying the DNA 1000 chip on the Bioanalyzer 2100 (Agilent, Santa Clara, USA). cDNA libraries were amplified and sequenced using the cBot and HiSeq2000 from Illumina (paired end; 2×100 bp). Sequence images were transformed with Illumina software BaseCaller to bcl files, which were demultiplexed to fastq files with CASAVA v1.8.2 (Illumina). Quality check was done via fastqc (v. 0.10.0, Babraham Institute, Cambridge, UK).
OBP and CSP re-annotation, SNP calling and mapping
The obtained fastq formatted Illumina reads were mapped to the Tribolium castaneum 3.0 official gene set using bowtie2 [64] with the “very-sensitive” presetting. The previously published CSP and OBP sequences [7, 17, 21, 65] were identified in this gene set with blastp [66] implemeted in bioperl [67]. Samtools mpileup (v0.1.18) [68] was used to check the RNAseq data for SNPs and indels. In a genome independent approach a de novo assembly was built. Quality filtering was performed with the NGSQC Toolkit (v2.3.1) [69] in three steps: 1) removal of reads containing ambiguous bases with AmbiguityFiltering.pl, all settings default; 2) trimming of bad quality bases from 3′ ends with TrimmingReads.pl, -q 28 -n 60; 3) removal of bad quality reads with IlluQC_PRLL.pl, N 5 -l 90 -s 24. Before the assembly the reads were digitally normalized using the normalize_by_kmer_coverage.pl script from trinity (release2013_08_14) [70] with these settings: --max_cov 50 --pairs_together. The assembly was performed with Trinity.pl, all settings default. Translations of open reading frames were extracted with transcripts_to_best_scoring_ORFs.pl. The preliminary re-annotation of the whole T. castaneum gene set (au3) was generated by the gene finder AUGUSTUS [71]. Alignments of RNA-Seq reads from libraries from several tissues, stages and conditions (e.g. embryo, larva head, larva body, early and late stage pupa, adult antenna, leg, head, body, stink glands, ovary) were incorporated. These data were produced mainly by the iBeetle consortium [72] and a separate publication is in preparation describing the re-annotation of the whole gene set of T. castaneum based on these RNA-Seq data, which can be viewed in a respective genome browser [73]. It contains a track with the au3 gene models as well as RNA-Seq coverage tracks of different stages including the data collected for this study. In total 1,624,983,955 reads were mapped against the genome with the alignment tool BLAT [74]. The read alignments were filtered, so that only alignments of reads that mapped uniquely to the genome and that showed a percent identity of at least 93% were kept. Paired reads were required to be aligned in the correct orientation and with a maximal genomic distance of 500,000 base pairs. Intron evidence was collected based on reads with a spliced alignment against the genome and evidence for transcription is taken from RNA-Seq covered regions. In an iterative process, the SNP calling, the de novo assembly, and the au2 gene set were used to manually curate the OBP and CSP sequences based on previously published annotations [7, 17, 21, 65]. The corresponding au3 gene models were replaced with these new candidate sequences and the resulting modified au3 gene set was used to remap the RNAseq data with bowtie2 using the ‘very-sensitive’ presetting. Finally all sequences were searched for signal peptides using the SignalP4.1 server [75] and browsed for conserved functional domains [76].
Tribolium castaneumexpression profiling
The mapped reads of the re-annotated OBPs and CSPs in the particular tissue or sex sample were counted with samtools [68]. To normalize the count numbers RPKM values were calculated and plotted as log2 [RPKM + 1] (Additional file 1: Table S1). The values were visualized using the matrix2png interface (version 1.2.1; [77]) and the figures were composed with inkscape [78]. Male and female reads from the sequenced tissues were pooled and considered as biological replicates. Statistical analysis of the data was performed in R [79] using the DESeq package (version1.12.0) [80] from bioconductor [81]. All sequenced tissues were compared to body as reference. Significant differentially expressed genes (false discovery rate < 0.05) are marked with asterisks. For the intersex comparison the two male and three female replicates of antenna were treated the same way.
Phylogenetic analysis and interspecies comparison
We compared our sequences on protein level with data from D. melanogaster and the malaria mosquito Anopheles gambiae obtained from Vieira and Rozas 2011 [17]. After subtraction of the signal peptide (SignalP4.1) [75], the sequences were aligned using MAFFT v7.040b [82] as described [17] and the tree was constructed using RAxML version 7.8.6. [83] with the LG substitution model in the case of the CSPs or the VT substitution model for the OBPs and GAMMA correction. Node support was assessed with 100 rapid bootstrap replicates. The relative expression levels were calculated as log2 fold changes of antenna/body and palp (mouthpart)/body. For T. castaneum, log2FC data from inner species comparison were used. The D. melanogaster data set was downloaded from EMBL gene expression atlas [84] originally published in Farhadian et al. 2012 [43] and the An. gambiae data were obtained from Pitts et al. 2011 [42]. The phylogenetic tree was visualized by iTOL [85] and descriptions were added using inkscape [78]. Since the absolute expression levels of the different candidates are lost in the depiction of the fold changes, we provide them in Additional file 2: Figure S1. Please note that the methods used to obtain the different expression data (RNA-seq and microarray) are not directly comparable. Therefore, Additional file 2: Figure S1 can just give an impression on more or less abundant transcripts.
Cloning of selected OBPs and CSPs open reading frames
Manually separated heads were ground in liquid nitrogen, and total RNA was extracted using the TRIZOL reagent (life technologies, Carlsbad, USA). Messenger RNA was purified with the Dynabeads purification kit (life technologies, Carlsbad, USA) and cDNA was synthesized using the Super-Script first-strand synthesis system (life technologies, Carlsbad, USA). Hotstart Taq DNA polymerase (Qiagen, Venlo, Netherlands) was used to amplify individual transcripts. Finally the products were cloned into PCR2.1 vector (life technologies, Carlsbad, USA) and verified by sequencing. Most primers were designed to bind within the UTRs to not bias start and stop codons and are summarized in Additional file 1: Table S1.
MALDI-TOF MS
For identification of OBPs and CSPs on protein level, about 400 antennae per sample were manually separated and homogenized in 200 μl milliQ water containing 0.1% trifluoroacetic acid (Sigma-Aldrich, St. Louis, USA) with a tube fitting pestle. To get rid of the debris, the samples were centrifuged and 150 μl supernatant was used further. To break down the secondary structure, the disulfide bridges were reduced and simultaneously the cysteine-derived thiol groups alkylated with 10 μl 100 mM tris (2-carboxyethyl) phosphine hydrochloride (Chemos GmbH, Regenstauf, Germany), 10 μl 200 mM 2-vinylpyridine (in 30% acetonitrile, Sigma-Aldrich) and 26 μl 8 M guanidine hydrochloride (Sigma-Aldrich) for 8 min at 35°C, followed by additional incubation for 30 min. at pH 8 after adding 11 μl 1 M ammonium bicarbonate (Sigma-Aldrich). The sample was loaded on a VIVASPIN 500 VS011 Ultrafiltration unit (5000 MWCO, Sartorius, Goettingen, Germany) and centrifuged for 10 min followed by two washing steps with 200 μl milliQ. For storage at -20°C over night the remaining 50 μl sample was mixed with 100 μl milliQ and 50 μl acetonitrile. After 30 min of centrifugation, 100 μl milliQ, 50 μl 50 mM Ammonium bicarbonate and 20 μl acetonitrile were added and debris was removed by additional centrifugation. Digestion took place in the remaining volume over the membrane by adding 0.11 μg sequencing-grade modified trypsin (Promega, Fitchburg, USA) and the resulting peptides were eluted. The mixture was analyzed by nanoLC (PepMap100 C-18 RP nanocolumn and UltiMate 3000 liquid chromatography system; Dionex, Sunnyvale, USA) and automated MSMS (4800 Proteomics Analyzer MDS¸ AB Sciex, Framingham, USA). MSMS data were searched against the au2 gene set (http://bioinf.uni-greifswald.de/gb2/gbrowse/tcas4/) using Mascot embedded into GPS explorer software (AB Sciex). Identified proteins, their scores, and Pfam predictions are provided in Additional file 3: Table S2.
Results and discussion
Re-annotation and re-naming of TriboliumCSPs and OBPs
In the past, several authors published sequences of Tribolium CSPs and OBPs based on computational predictions [7, 17, 21, 65] resulting in different conflicting annotations and designation. We revised the originally described 20 CSPs and 50 OBPs using transcriptome analysis of different tissues including antennae and mouthparts. Subsequently we applied a new nomenclature to prevent confusion and to provide a unique and distinguishable nomenclature following the one used for Drosophila OBPs [22]. We used the prefaces CSP, and OBP to reflect the fact that a gene is a member of one of these protein families. This is followed by a number reflecting the chromosomal location and a letter that conveys its relative position on the chromosome (Figure 1A). Thus, the new name OBP9B refers to the second OBP on the ninth chromosome. A comparative list putting all previous names in relation can be found in the Additional file 1: Table S1.
Chromosomal localization and chemosensory expression profile of Tribolium CSPs and OBPs. (A) Chromosomal localization of CSPs (magenta), classic OBPs (black), ABPII (grey), C-OBPs (green) and C + OBP (blue), based on Georgia GA-2 strain genome assembly 3.0 [7]. The arrowheads indicate the orientation of the genes from 5′►3′. Genes encoding for more than the six highly conserved cysteines (or four in case of C-OBP) are labeled with an asterisk (*). (B) Heatmap showing the absolute expression level of the OBPs/CSPs as log2 (RPKM + 1) in different tissues (adult antennae, head (missing antennae but including mouth parts), mouthparts, legs, body, as well as larval head and body). The candidates are blotted according to their chromosomal localization, horizontal brackets above indicate clustering in the genome. A black dot in the first row labeled ‘MALDI’ indicates that at least one unique tryptic fragment of the particular candidate was identified in an antennae sample on protein level. The expression levels are represented by a log2 greyscale with high expression levels (216 RPKM) labelled black. The asterisks mark statistically significant differtially expressed genes compared to body. The red asterisks represent up- and the blue down-regulation (p-values are * < 0.05; ** < 0.01; *** < 0.001).
We detected reads corresponding to all previously described Tribolium CSPs and OBPs except TcOBP2A. Accordingly we confirmed or corrected the predicted open reading frames of all 20 CSPs and 49 OBPs. In case of low abundant transcripts with poor read coverage in our samples we used additional data obtained from embryo and pupa to support the re-annotation (iBeetle genome browser) [73]. The comparison of the latest genome based annotation [17] with our transcriptome based re-annotation revealed differences mainly based on wrongly predicted intron-exon boundaries. The identified discrepancies to previous annotations in the OBPs and one CSP did not cause severe differences in the phylogenetic relationship. However, they partially affect in addition to the intron-exon boundaries also the start or stop codons, which could impair cloning efforts for further investigations. In addition, wrong indels can cause differences in three-dimensional modeling of OBPs and by this also affect predictions in respect to potential ligands. Therefore, we point out clearly which annotations of previously identified OBPs and CSPs needed to be changed: Due to an unpredicted intron in the 5′UTRs of TcCSP2A, TcOBP0A and TcOBP5F the start codons had to be changed. In case of TcOBP5E, TcOBP6A, TcOBP6E, TcOBP6F, TcOBP8A, TcOBP7G and TcOBP7H wrong or un-predicted introns led to insertions or deletions not affecting the highly conserved cysteine pattern. In TcOBP4A, TcOBP4C, TcOBP4E, TcOBP4F, TcOBP5C, TcOBP5D and TcOBP5H an incorrect last intron caused differences in the C-terminal region, that was supposed to be involved in ligand binding and release [86, 87]. Additionally we were able to revive previously wrongly annotated pseudogenes. Formerly termed OBP49P is an intact gene with some characteristics of an OBP and is now called TcOBP7C. OBP50P [17] turned out to correspond to the already described OBP-C08 [7] and is now called TcOBP7K. The originally termed CSP21P, which is derived from a duplication of the first exon of TcCSP7G, is expressed at low levels and now called TcCSP7H, which either represents an expressed pseudogene as observed in Nasonia[88] or is a truncated CSP. Because of its high sequence similarity to TcCSP7G we were only able to map unique reads to the 3′UTR. Therefore, expression of TcCSP7H is not indicated in our figures. To confirm the new annotation we checked the resulting protein sequences for common characteristics of CSP- or OBP-like signal peptide, conserved domains, size and cysteine composition [89] and we cloned the whole open reading frame of 33 of the 70 candidates from cDNA. All newly annotated proteins contain a predicted N terminal signalpeptide with an length of 15 to 22 AA (SignalP 4.1 [75]). According to the conserved domain database [76], all re-annotated CSPs and OBPs (except the C + OBP, OBP5E) are members of the OS-D respectively PBP_GOBP superfamily (Additional file 1: Table S1).
The re-annotated CSPs of Tribolium have an average size of 128 AA (110 AA mature CSP), they range from the smallest, TcCSP7C, with just 99 AA to the longest, TcCSP7E, with 251 AA. On average, Tribolium CSPs are within the size range of other species [12]. The cysteine formula of the Tribolium CSPs follows the highly conserved pattern with four cysteines arranged by an exact spacing of C1X6C2X18C3X2C4 [17]. The only exception is the pseudo/truncated gene TcCSP7H that stops after C2.
The OBPs are slightly longer and vary from 106 up to 320 AA with an average size of 143 AA (125 for the mature OBP), which is similar to other insects such as D. melanogaster (117 up to 245 AA) or An. gambiae (107 up to 356 AA [17]). Most of the T. castaneum classical OBPs (including ABPIIs) show a conserved cysteine pattern (C1X24-29C2X3C3X22-43C4X8-10C5X8C6) comparable to D. melanogaster[22] and other insects. The only exceptions are the C + OBP (TcOBP5E, 241 AA) and the non-clustered OBPs TcOBP5A, TcOBP7C, and TcOBP8B (Figure 1A), which differ in C spacing, are of unusual length (TcOBP5E, 241AA; TcOBP5A, 176 AA; TcOBP7C 320 AA; TcOBP8B, 200 AA), and are phylogenetically close to C + OBPs. The one typical C + OBP, TcOBP5E, has an expanded N and C terminus containing six additional cysteines, whereas TcOBP7C has an expanded N terminus, TcOBP8B extra AAs between C1 and C2, TcOBP5A between C1 and C2 plus between C4 and C5. TcOBP5A, TcOBP8B, and TcOBP5E have extra cysteines 17–19 AA before C1 and nine AA after C6. The OBPs TcOBP5B, TcOBP5C, TcOBP5D, TcOBP6E, TcOBP6G, TcOBP7C, and TcOBP8A contain also at least one additional cysteine. TcOBP6G and the ABPIIs (TcOBP5B, TcOBP5C, TcOBP5D) have a conserved additional cysteine seven AA after C3. Despite the increased amount of cysteines only TcOBP5E carries the typical proline residue following C6 (C6b) [90, 91]. The C-OBPs show a conserved cysteine pattern, with only four cysteines, lacking C2 and C5 (C1X18-30C3X37-39C4X16-17C6). We can conclude that all re-annotated CSPs (except CSP7H) and OBPs fulfill the rigid criteria previously defined based on other species [22, 89].
Expression profile of the CSPs and OBPs in Tribolium castaneum
As several CSPs and OBPs are supposed to be involved in olfaction, we comparatively analyzed the expression of these genes in the main chemosensory tissues antennae and mouthparts (here defined as the piece of the head capsule anterior to the antennae) plus in heads (the whole head capsule excluding the antennae), legs, and bodies (excluding head and legs) of males or females, respectively. To get some first insights into expression differences between larval and adult stages, we also sequenced heads including antennae and bodies (without head) of last instar larvae. The results as log2 RPKM are represented as heat-map in Figure 1B.
The expression of the majority of the CSPs is detected in a wide variety of tissues. Transcripts of only five of the 20 CSPs are significantly enriched in antennae (TcCSP7A, TcCSP7G, TcCSP7J, TcCSP7K, and TcCSP7R) and six in the mouthparts (TcCSP7A, TcCSP7B, TcCSP7G, TcCSP7J, TcCSP7K, and TcCSP7R). However, only the expression of TcCSP7A and TcCSP7G is restricted to the main olfactory tissue. TcCSP7I and TcCSP7M are exclusively expressed in the body. Six of the CSPs showed no or only poor expression in our tissue samples, however, we found them expressed at other developmental stages by searching the iBeetle genome browser [73]. TcCSP7P and TcCSP7Q are expressed in embryo and pupa, TcCSP3A mainly in embryo, TcCSP7N in embryo and larva, and TcCSP7O only in larva.
In contrast to the CSPs, the expression of the OBPs is more restricted to the main chemosensory tissues (antennae and mouthparts). All eight ABPIIs are highly expressed in the antennae. With the exception of TcOBP5C, all of them are also significantly enriched in mouthparts indicating an involvement of this subgroup in chemo-reception (Figure 1B). 15 of the 20 classic OBPs are significantly enriched in antennae and mouthparts (Figure 1B, TcOBP0A, TcOBP4B, TcOBP4C, TcOBP4D, TcOBP4E, TcOBP4F, TcOBP6B, TcOBP6C, TcOBP6D, TcOBP6E, TcOBP6F, TcOBP6G, TcOBP7C, TcOBP8A, TcOBP8B), whereas three are enriched only in the mouthparts (TcOBP4A, TcOBP4J, TcOBP7D). Only three of the classic OBPs are evenly expressed in all tissues. Interestingly, these are the non-clustered ones (TcOBP5A, TcOBP6A, TcOBP7D). The C + OBP (TcOBP5E) is enriched in mouthparts and in antennae. Most of the 21 C-OBPs are expressed in all tissues similar to the majority of the CSPs, only five are significantly enriched in antennae and mouthparts compared to body with three of them also highly abundant in head or leg (TcOBP4G, TcOBP4I, TcOBP7E). Thus, there are just two C-OBPs (TcOBP7L and TcOBP9C) most likely exclusively involved in chemosensory processing. Ten C-OBPs are significantly down-regulated in the antennae compared to the body: TcOBP3A, TcOBP7B, TcOBP7G, TcOBP7H, TcOBP7I, TcOBP7J, TcOBP10A, TcOBP10B, TcOBP10C, and TcOBP10D. TcOBP2A expression was not detected at all, TcOBP7F is expressed during metamorphosis and TcOBP7B is mainly active during embryogenesis as well as metamorphosis.
Statistical analysis of the two male and three female antennal samples did not show any significant difference, due to the low abundance of potential candidates and the relative high dispersion of the samples (Figure 2; Additional file 4: Figure S2, Additional file 5: Figure S3, Additional file 6: Figure S4 and Additional file 7: Figure S5). Nevertheless, TcOBP7A, TcOBP7K, and TcOBP7M were more than five-fold overexpressed in male antennae and could be interesting for further investigation (Figure 2). The fact that we found no major and significant differences between male and female is consistent with anatomical data from the antennal lobe were no sexual dimorphism was found [92], and the antennal morphology from a related species, Tribolium brevicornis, were both sexes are anatomically similar [93].
Comparison of expression levels of CSPs and OBPs in male and female antennae, average values based on two male and three female antennal samples. Scatter plot of the RPKM values of the CSPs (in pink) and OBPs (classic in black, ABPII in grey, C-OBP in green, C + OBP in blue). The dotted lines represent a fivefold difference.
The comparison of transcriptome data of adult antennae and larval heads revealed differences in the expression of five ABPIIs (TcOBP5C, TcOBP5F, TcOBP5G, TcOBP5H, and TcOBP9A), one classic OBP (TcOBP4D) and five C-OBPs (TcOBP3A, TcOBP4G, TcOBP4I, TcOBP7I, and TcOBP9C). Most of these transcripts were present in adult antennae or mouthparts but were absent in the larval head (Figure 1B) reflecting the reduced larval olfactory system which also corresponds to the lower amount of expressed odorant receptors previously described (41 in larvae compared to 111 in adults) [94].
In a previous study regarding the stink glands of T. castaneum[95], TcOBP8A, TcOBP6B, TcCSP7P and TcCSP7R were identified to be transcriptionally enriched in the prothoracic glands compared to general anterior abdominal tissue. Interestingly TcOBP8A and TcOBP6B were also enriched in antennae and mouthparts, whereas TcCSP7P was only detected in embryo and pupa. It seems that in Tribolium these OBPs are not only involved in reception of odorants/pheromones but also in production or release of such semiochemicals, as postulated for some lepidoptera [26, 49].
To relate our transcriptome data to protein detection in the adult antenna, we additionally performed MALDI-TOF-TOF MS with antennal extracts. We were able to identify fingerprints of 14 of the 70 candidates in antennae on protein level (Figure 1B). Thus, we found at least one tryptic fragment with an ion score above 50 that maps uniquely to the AA sequence of one of the CSPs or OBPs (Additional file 3, Table S2). We identified three highly expressed CSPs (TcCSP7A, TcCSP7G, TcCSP7R), the three highest expressed classic OBPs (TcOBP4C, TcOBP4D, TcOBP4E), four of the highly expressed ABPII subclass (TcOBP5C, TcOBP5D, TcOBP5H, TcOBP9B), and four C-OBPs (TcOBP7E, TcOBP7J, TcOBP10A, TcOBP10B). All identified tryptic fragments belong to genes that are transcribed in the antennae, therefore confirming their expression also on a protein level.
Phylogenetic considerations in respect to the expression of the CSPs and OBPs
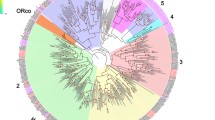
The majority of the 50 OBPs and 20 CSPS of T. castaneum are arranged in clusters like in other insects e.g. D. melanogaster[22], An. gambiae[96], the honey bee Apis mellifera[21] and silk moth Bombyx mori[97]. The CSPs are organized in arrays of two, six and ten genes on the seventh chromosome, only two (TcCSP2A, TcCSP3A) are non-clustered and located on chromosome 2 and 3. Most of the classic OBPs are arranged in two large arrays on chromosome 4 and 6, only six are interspersed (TcOBP5A, TcOBP6A, TcOBP7C, TcOBP7D, TcOBP8A and TcOBP8B). Six of the eight ABPIIs are located on chromosome 5 with three genes per cluster, the remaining two are close together on Chromosome 9. Nine of the 21 C-OBPs are located in a cluster on chromosome 7, close to the interspersed classic OBP TcOBP7D, that is phylogenetically the closest relative classic OBP to all C-OBPs. Additional three C-OBPs form a cluster on chromosome 4, four on chromosome 10 and the remaining 5 C-OBPs are interspersed on chromosome 2, 7 and 9. The only C + OBP (TcOBP5E) is located next to the second ABPII cluster on chromosome 5, but is phylogenetically unrelated to this group. All other OBPs carrying an additional cysteine are randomly distributed over the genome. The presence of clusters of phylogenetically related genes in all investigated insects can be explained by their origin from gene duplication events within the respective lineage but the fact that the clusters are conserved within different Drosophilidae indicates some constraints that stabilize the clusters [98]. One possible explanation for the maintaining of the clusters is the sharing of regulative elements [99], however, our expression data do not support this theory since genes from the same cluster (Figure 1B, indicated by horizontal brackets) show partially unrelated expression. Most likely more sophisticated methods are needed to understand the complex interplay of regulative elements within a cluster as recently shown for regulatory elements of odorant receptors in Drosophila[100] and their distribution within clusters. An interspecies comparison between T. castaneum, D. melanogaster, and An. gambiae regarding the expression level in a phylogenetic context revealed that some expression features found in T. castaneum are conserved between these species. The majority of the CSPs of all species are expressed in all tissues (Figure 3A). The classic OBPs in the branch holding genes of all three species are mainly enriched in antennae and/or mouthparts (Figure 3B). Only three – namely DmOBP22a, DmOBP56f, DmOBP51a – are clearly underrepresented in the main chemosensory tissues. Also the antennal expression of T. castaneum ABPIIs is consistent in the other species (Figure 3B). All members of this subgroup except AgOBP18 are enriched in antennae and the highest expressed OBPs within each species belong to this group.
Mid-point rooted phylogenetic tree of CSP (A) and OBP (B) sequences from Tribolium castaneum (green branches), Drosophila melanogaster (red branches), and Anopheles gambiae (blue branches). Outer rings represent the expression in antennae and ‘mouthparts’ (Tribolium: palps, mandible, labrum and labium; Drosophila: palp and proboscis; Anopheles: maxillary palp) as log2 fold change compared to body corresponding to the scale in the right middle. The scale bars within the trees represent 1 amino acid substitution per site. Inner ring in B indicates the phylogenetic subclass (classic in black, ABPII in grey, C-OBP in green, C + OBP in blue, D7 in yellow, Dimer in red). Numbers on branches show values of 100 times replication bootstrap analysis higher than 70.
Multiple alignments of CSPs (A), OBPs (B), and C-OBPs including TcOBP7D (C) made with ClustalW2[94]and visualized with Jalview 2.0.1[95]. OBP8B was manually adapted to fit the conserved cysteine pattern. The N-terminal regions of OBP7C, 8B and 5E are truncated, as well as a portion between C1 and C2 of OBP5A and OBP7C, and the C-terminus of OBP5E and OBP6A (the number of deleted AA is indicated, respectively). Cysteines are indicated by red frames. CSPs are indicated in pink, classic OBPs in black, ABPIIs in grey, C-OBPs in green, and the C + OBP in blue.
The comparison of the C-OBPs of T. castaneum, An. gambiae, and D. melanogaster revealed that they are polyphyletic (Figure 3B) as previously shown by Vieira and Rozas [17]. The phylogenetic analysis as well as the chromosomal clustering indicates that in T. castaneum this large expanded group is together with the classic OBP TcOBP7D most likely derived from a common ancestor. This is similar to the situation in A. mellifera, where a monophyletic group of C-OBPs (AmOBP14 to AmOBP21) clusters together with the classic OBP AmOBP13 both on the genomic localization and on the phylogenetic level [21]. However, even though the C-OBPs of different species are polyphyletic in their origin, they are in general highly and equally expressed in all tissues indicating a broad function. The loss of a disulfide bridge might increase their binding flexibility to serve different binding tasks [10, 20, 101]. The C-OBPs (21 in T. castaneum, four in D. melanogaster) actually represent similarly to the CSPs (20 in T. castaneum, four in D. melanogaster, eight in An. gambiae) a large expansion in T. castaneum and are mostly not antennae- or mouthpart-specifically expressed. Therefore, these proteins might not be mainly involved in chemosensory detection but might have additional roles such as detoxification which has been discussed for D. sechelia[102].
Conclusion
Our T. castaneum expression analysis revealed expression of most CSPs and C-OBPs in various body parts, whereas expression of classical OBPs and ABPIIs is mainly restricted to the antennae and mouthparts. These data are consistent with previous observations in different insects [48], like A. meliffera[21, 65], An. gambiae[42], B. mori[103] and the large black chafer Holotrichia parallela[104]. Systematic OBP knockdowns in D. melanogaster show their necessity for correct olfactory behavioral responses and indicate a combinatorial OBP-dependent odorant recognition [38]. Our comparative expression data suggest that within the classic OBPs, especially the ABPII subgroup has a specific role in olfaction, since all members of T. castaneum, An. gambiae, and D. melanogaster are highly expressed and enriched in the antennae. Moreover, this group contains some of the most prominent OBPs such as D. melanogaster LUSH involved in pheromone detection [35, 41], An. gambiae AgOBP4 that forms cooperative heteromers with other OBPs [105], and AgOBP1 that mediates indole detection to find blood meals [39] and is co-expressed with other ABPIIs (AgOBP3, AgOBP4, AgOBP19) [106].
Most OBPs of T. castaneum are arranged in clusters in the genome. The only exceptions are TcOBP5A, TcOBP6A, TcOBP7C, TcOBP7D, TcOBP8A, and TcOBP8B. Intriguingly three of them (TcOBP5A, TcOBP6A, TcOBP7D) show an atypical, ubiquitous expression and four differ massively from the average size of 143 AA: TcOBP8A 106 AA; TcOBP5A 176 AA, TcOBP8B 200 AA, and TcOBP7C 320 AA (Figure 4). Interestingly, a bootstrap value of 100 indicates orthology of TcOBP5A with a single widely expressed OBP in An. gambiae (AgOBP80) and D. melanogaster (DmOBP73a) (Figure 3), which seem to have also single orthologues in B. mori, the pea aphid Acyrthosiphon pisum, and the body louse Pediculus humanus[17].
In contrast to the ABPIIs and classic OBPs, the CSPs and C-OBPs show a more broad expression indicating a more versatile function in transport of hydrophobic chemicals involved in various processes. In T. castaneum, several functions besides semiochemical reception are implicated: TcCSP7P and TcCSP7R are highly enriched in odoriferous glands [95] and may be involved in the secretion of semiochemicals or defensive products; TcOBP10B and TcCSP7D are up-regulated after cry toxin exposure [107] indicating a function in detoxification or the innate immune system of T. castaneum. TcOBP7F is transferred during copulation via seminal fluids [108] similar to the yellow fever mosquito Aedes aegyti[109] to potentially mark fertilized eggs, as also described for Helicoverpa moths [50]. Many more functions of CSPs in insects have sporadically been described, such as involvement in limb regeneration in the American cockroach Periplaneta americana[53], presence in the female reproductive organs of the migratory locust Locusta migratoria[110], involvement in embryonic integument formation in A. mellifera[51], response to an insecticide in the silverleaf whitefly Bemisia tabaci[111] and B. mori[112], detergent like function in the proboscis of two Helicoverpa species [113]. Taken together, some CSPs seem to participate in chemoreception, however, most of them might have more general functions involved in the release of semiochemicals [26], development [51], reproduction [50, 108, 109, 114], food intake [113], and in the drug/immune response [52, 107, 111, 112].
Availability of supporting data
The complete transcriptomics dataset including all relevant parameters has been deposited to the National Center for Biotechnology Information (NCBI) database repository ‘Gene Expression Omnibus’ (GEO accession number: GSE63162) [115].
References
Sokoloff A: The Genetics of Tribolium and Related Species. 1966, New York: Academic Press, 1:
Bucher G, Scholten J, Klingler M: Parental RNAi in Tribolium (Coleoptera). Curr Biol CB. 2002, 12: R85-R86. 10.1016/S0960-9822(02)00666-8.
Tomoyasu Y, Denell RE: Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev Genes Evol. 2004, 214: 575-578. 10.1007/s00427-004-0434-0.
Trauner J, Schinko J, Lorenzen MD, Shippy TD, Wimmer EA, Beeman RW, Klingler M, Bucher G, Brown SJ: Large-scale insertional mutagenesis of a coleopteran stored grain pest, the red flour beetle Tribolium castaneum, identifies embryonic lethal mutations and enhancer traps. BMC Biol. 2009, 7: 73-10.1186/1741-7007-7-73.
Schinko JB, Weber M, Viktorinova I, Kiupakis A, Averof M, Klingler M, Wimmer EA, Bucher G: Functionality of the GAL4/UAS system in Tribolium requires the use of endogenous core promoters. BMC Dev Biol. 2010, 10: 53-10.1186/1471-213X-10-53.
Schinko JB, Hillebrand K, Bucher G: Heat shock-mediated misexpression of genes in the beetle Tribolium castaneum. Dev Genes Evol. 2012, 222: 287-298. 10.1007/s00427-012-0412-x.
Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, Beeman RW, Gibbs R, Beeman RW, Brown SJ, Bucher G, Friedrich M, Grimmelikhuijzen CJP, Klingler M, Lorenzen M, Richards S, Roth S, Schröder R, Tautz D, Zdobnov EM, Muzny D, Gibbs RA, Weinstock GM, Attaway T, Bell S, Buhay CJ, Chandrabose MN, Chavez D, Clerk-Blankenburg KP, Cree A, Dao M, et al: The genome of the model beetle and pest Tribolium castaneum. Nature. 2008, 452: 949-955. 10.1038/nature06784.
Kim HS, Murphy T, Xia J, Caragea D, Park Y, Beeman RW, Lorenzen MD, Butcher S, Manak JR, Brown SJ: BeetleBase in 2010: revisions to provide comprehensive genomic information for Tribolium castaneum. Nucleic Acids Res. 2010, 38 (Database issue): D437-D442.
de Santis F, François M-C, Merlin C, Pelletier J, Maïbèche-Coisné M, Conti E, Jacquin-Joly E: Molecular Cloning and in Situ Expression Patterns of Two New Pheromone-Binding Proteins from the Corn Stemborer Sesamia nonagrioides. J Chem Ecol. 2006, 32: 1703-1717. 10.1007/s10886-006-9103-2.
Angeli S, Ceron F, Scaloni A, Monti M, Monteforti G, Minnocci A, Petacchi R, Pelosi P: Purification, structural characterization, cloning and immunocytochemical localization of chemoreception proteins from Schistocerca gregaria. Eur J Biochem. 1999, 262: 745-754. 10.1046/j.1432-1327.1999.00438.x.
Pelosi P, Zhou J-J, Ban LP, Calvello M: Soluble proteins in insect chemical communication. Cell Mol Life Sci CMLS. 2006, 63: 1658-1676. 10.1007/s00018-005-5607-0.
Wanner KW, Willis LG, Theilmann DA, Isman MB, Feng Q, Plettner E: Analysis of the Insect OS-D-Like Gene Family. J Chem Ecol. 2004, 30: 889-911.
Liu R, He X, Lehane S, Lehane M, Hertz-Fowler C, Berriman M, Field LM, Zhou J-J: Expression of chemosensory proteins in the tsetse fly Glossina morsitans morsitans is related to female host-seeking behaviour. Insect Mol Biol. 2012, 21: 41-48. 10.1111/j.1365-2583.2011.01114.x.
Leal WS: Odorant Reception in Insects: Roles of Receptors, Binding Proteins, and Degrading Enzymes. Annu Rev Entomol. 2013, 58: 373-391. 10.1146/annurev-ento-120811-153635.
Steinbrecht RA: Odorant-binding proteins: expression and function. Ann N Y Acad Sci. 1998, 855: 323-332. 10.1111/j.1749-6632.1998.tb10591.x.
Vogt RG, Riddiford LM: Pheromone binding and inactivation by moth antennae. Nature. 1981, 293: 161-163. 10.1038/293161a0.
Vieira FG, Rozas J: Comparative Genomics of the Odorant-Binding and Chemosensory Protein Gene Families across the Arthropoda: Origin and Evolutionary History of the Chemosensory System. Genome Biol Evol. 2011, 3: 476-490. 10.1093/gbe/evr033.
Sandler BH, Nikonova L, Leal WS, Clardy J: Sexual attraction in the silkworm moth: structure of the pheromone-binding-protein-bombykol complex. Chem Biol. 2000, 7: 143-151. 10.1016/S1074-5521(00)00078-8.
Briand L, Swasdipan N, Nespoulous C, Bézirard V, Blon F, Huet J-C, Ebert P, Penollet J-C: Characterization of a chemosensory protein (ASP3c) from honeybee (Apis mellifera L.) as a brood pheromone carrier. Eur J Biochem FEBS. 2002, 269: 4586-4596. 10.1046/j.1432-1033.2002.03156.x.
Scaloni A, Monti M, Angeli S, Pelosi P: Structural analysis and disulfide-bridge pairing of two odorant-binding proteins from Bombyx mori. Biochem Biophys Res Commun. 1999, 266: 386-391. 10.1006/bbrc.1999.1791.
Foret S, Maleszka R: Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera). Genome Res. 2006, 16: 1404-1413. 10.1101/gr.5075706.
Hekmat-Scafe DS, Scafe CR, McKinney AJ, Tanouye MA: Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 2002, 12: 1357-1369. 10.1101/gr.239402.
Spinelli S, Lagarde A, Iovinella I, Legrand P, Tegoni M, Pelosi P, Cambillau C: Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules. Insect Biochem Mol Biol. 2012, 42: 41-50. 10.1016/j.ibmb.2011.10.005.
Gu S-H, Wang S-Y, Zhang X-Y, Ji P, Liu J-T, Wang G-R, Wu K-M, Guo Y-Y, Zhou J-J, Zhang Y-J: Functional Characterizations of Chemosensory Proteins of the Alfalfa Plant Bug Adelphocoris lineolatus Indicate Their Involvement in Host Recognition. PLoS One. 2012, 7: e42871-10.1371/journal.pone.0042871.
Ozaki M: Ant Nestmate and Non-Nestmate Discrimination by a Chemosensory Sensillum. Science. 2005, 309: 311-314. 10.1126/science.1105244.
Jacquin-Joly E, Vogt RG, François M-C, Meillour PN-L: Functional and Expression Pattern Analysis of Chemosensory Proteins Expressed in Antennae and Pheromonal Gland of Mamestra brassicae. Chem Senses. 2001, 26: 833-844. 10.1093/chemse/26.7.833.
Andersson MN, Grosse-Wilde E, Keeling CI, Bengtsson JM, Yuen MM, Li M, Hillbur Y, Bohlmann J, Hansson BS, Schlyter F: Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae). BMC Genomics. 2013, 14: 198-10.1186/1471-2164-14-198.
González D, Zhao Q, McMahan C, Velasquez D, Haskins WE, Sponsel V, Cassill A, Renthal R: The major antennal chemosensory protein of red imported fire ant workers. Insect Mol Biol. 2009, 18: 395-404. 10.1111/j.1365-2583.2009.00883.x.
Liu X, Luo Q, Zhong G, Rizwan-Ul-Haq M, Hu M: Molecular characterization and expression pattern of four chemosensory proteins from diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). J Biochem (Tokyo). 2010, 148: 189-200. 10.1093/jb/mvq050.
Forstner M, Breer H, Krieger J: A receptor and binding protein interplay in the detection of a distinct pheromone component in the silkmoth Antheraea polyphemus. Int J Biol Sci. 2009, 5: 745-757.
Grosse-Wilde E, Gohl T, Bouché E, Breer H, Krieger J: Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur J Neurosci. 2007, 25: 2364-2373. 10.1111/j.1460-9568.2007.05512.x.
Grosse-Wilde E, Svatos A, Krieger J: A pheromone-binding protein mediates the bombykol-induced activation of a pheromone receptor in vitro. Chem Senses. 2006, 31: 547-555. 10.1093/chemse/bjj059.
Hallem EA, Ho MG, Carlson JR: The molecular basis of odor coding in the Drosophila antenna. Cell. 2004, 117: 965-979. 10.1016/j.cell.2004.05.012.
Syed Z, Ishida Y, Taylor K, Kimbrell DA, Leal WS: Pheromone reception in fruit flies expressing a moth’s odorant receptor. Proc Natl Acad Sci U S A. 2006, 103: 16538-16543. 10.1073/pnas.0607874103.
Xu P, Atkinson R, Jones DNM, Smith DP: Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron. 2005, 45: 193-200. 10.1016/j.neuron.2004.12.031.
Arya GH, Weber AL, Wang P, Magwire MM, Negron YLS, Mackay TFC, Anholt RRH: Natural Variation, Functional Pleiotropy and Transcriptional Contexts of Odorant Binding Protein Genes in Drosophila melanogaster. Genetics. 2010, 186: 1475-1485. 10.1534/genetics.110.123166.
Krieger MJB, Ross KG: Molecular evolutionary analyses of the odorant-binding protein gene Gp-9 in fire ants and other Solenopsis species. Mol Biol Evol. 2005, 22: 2090-2103. 10.1093/molbev/msi203.
Swarup S, Williams TI, Anholt RRH: Functional dissection of Odorant binding protein genes in Drosophila melanogaster. Genes Brain Behav. 2011, 10: 648-657. 10.1111/j.1601-183X.2011.00704.x.
Biessmann H, Andronopoulou E, Biessmann MR, Douris V, Dimitratos SD, Eliopoulos E, Guerin PM, Iatrou K, Justice RW, Kröber T, Marinotti O, Tsitoura P, Woods DF, Walter MF: The Anopheles gambiae Odorant Binding Protein 1 (AgamOBP1) Mediates Indole Recognition in the Antennae of Female Mosquitoes. PLoS One. 2010, 5: e9471-10.1371/journal.pone.0009471.
Pelletier J, Guidolin A, Syed Z, Cornel AJ, Leal WS: Knockdown of a Mosquito Odorant-binding Protein Involved in the Sensitive Detection of Oviposition Attractants. J Chem Ecol. 2010, 36: 245-248. 10.1007/s10886-010-9762-x.
Gomez-Diaz C, Reina JH, Cambillau C, Benton R: Ligands for Pheromone-Sensing Neurons Are Not Conformationally Activated Odorant Binding Proteins. PLoS Biol. 2013, 11: e1001546-10.1371/journal.pbio.1001546.
Pitts RJ, Rinker DC, Jones PL, Rokas A, Zwiebel LJ: Transcriptome profiling of chemosensory appendages in the malaria vector Anopheles gambiae reveals tissue- and sex-specific signatures of odor coding. BMC Genomics. 2011, 12: 271-10.1186/1471-2164-12-271.
Farhadian SF, Suárez-Fariñas M, Cho CE, Pellegrino M, Vosshall LB: Post-fasting olfactory, transcriptional, and feeding responses in Drosophila. Physiol Behav. 2012, 105: 544-553. 10.1016/j.physbeh.2011.09.007.
Pelletier J, Leal WS: Genome Analysis and Expression Patterns of Odorant-Binding Proteins from the Southern House Mosquito Culex pipiens quinquefasciatus. PLoS One. 2009, 4: e6237-10.1371/journal.pone.0006237.
Zhang Y-N, Jin J-Y, Jin R, Xia Y-H, Zhou J-J, Deng J-Y, Dong S-L: Differential Expression Patterns in Chemosensory and Non-Chemosensory Tissues of Putative Chemosensory Genes Identified by Transcriptome Analysis of Insect Pest the Purple Stem Borer Sesamia inferens (Walker). PLoS One. 2013, 8: e69715-10.1371/journal.pone.0069715.
Zheng W, Peng W, Zhu C, Zhang Q, Saccone G, Zhang H: Identification and Expression Profile Analysis of Odorant Binding Proteins in the Oriental Fruit Fly Bactrocera dorsalis. Int J Mol Sci. 2013, 14: 14936-14949. 10.3390/ijms140714936.
Gong D-P, Zhang H, Zhao P, Lin Y, Xia Q-Y, Xiang Z-H: Identification and expression pattern of the chemosensory protein gene family in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2007, 37: 266-277. 10.1016/j.ibmb.2006.11.012.
Zhou J-J, Kan Y, Antoniw J, Pickett JA, Field LM: Genome and EST Analyses and Expression of a Gene Family with Putative Functions in Insect Chemoreception. Chem Senses. 2006, 31: 453-465. 10.1093/chemse/bjj050.
Dani FR, Michelucci E, Francese S, Mastrobuoni G, Cappellozza S, Marca GL, Niccolini A, Felicioli A, Moneti G, Pelosi P: Odorant-Binding Proteins and Chemosensory Proteins in Pheromone Detection and Release in the Silkmoth Bombyx mori. Chem Senses. 2011, 36: 335-344. 10.1093/chemse/bjq137.
Sun Y-L, Huang L-Q, Pelosi P, Wang C-Z: Expression in Antennae and Reproductive Organs Suggests a Dual Role of an Odorant-Binding Protein in Two Sibling Helicoverpa Species. PLoS One. 2012, 7: e30040-10.1371/journal.pone.0030040.
Maleszka J, Forêt S, Saint R, Maleszka R: RNAi-induced phenotypes suggest a novel role for a chemosensory protein CSP5 in the development of embryonic integument in the honeybee (Apis mellifera). Dev Genes Evol. 2007, 217: 189-196. 10.1007/s00427-006-0127-y.
Levy F, Bulet P, Ehret-Sabatier L: Proteomic Analysis of the Systemic Immune Response of Drosophila. Mol Cell Proteomics. 2004, 3: 156-166.
Nomura A, Kawasaki K, Kubo T, Natori S: Purification and localization of p10, a novel protein that increases in nymphal regenerating legs of Periplaneta americana (American cockroach). Int J Dev Biol. 1992, 36: 391-398.
Schütz S, Weißbecker B, Schroth P, Schöning MJ: Linkage of Inanimate Structures to Biological Systems — Smart Materials in Biological Micro- and Nanosystems. Smart Mater. Edited by: Hoffmann K-H. 2001, Heidelberg: Springer Berlin, 149-157.
Di Pietrantonio F, Cannatà D, Benetti M, Verona E, Varriale A, Staiano M, D’Auria S: Detection of odorant molecules via surface acoustic wave biosensor array based on odorant-binding proteins. Biosens Bioelectron. 2013, 41: 328-334.
Schütz S, Weißbecker B, Koch UT, Hummel HE: Detection of volatiles released by diseased potato tubers using a biosensor on the basis of intact insect antennae. Biosens Bioelectron. 1999, 14: 221-228. 10.1016/S0956-5663(98)00092-X.
Johne AB, Weissbecker B, Schütz S: Approaching risk assessment of complex disease development in horse chestnut trees: a chemical ecologist’s perspective. J Appl Entomol. 2008, 132: 349-359. 10.1111/j.1439-0418.2008.01283.x.
Zhou J-J, Field LM, He XL: Insect Odorant-Binding Proteins: Do They Offer an Alternative Pest Control Strategy?. Outlooks Pest Manag. 2010, 21: 31-34. 10.1564/21feb08.
Pannure A, Mutthuraju GP, Imran S: The sense of smell in insects: a target for pest management? - A review. Curr Biot. 2012, 6: 399-419.
Abdel-Sattar E, Zaitoun AA, Farag MA, Gayed SHE, Harraz FMH: Chemical composition, insecticidal and insect repellent activity of Schinus molle L. leaf and fruit essential oils against Trogoderma granarium and Tribolium castaneum. Nat Prod Res. 2010, 24: 226-235. 10.1080/14786410802346223.
Andreev D, Kreitman M, Phillips TW, Beeman RW, ffrench-Constant RH: Multiple origins of cyclodiene insecticide resistance in Tribolium castaneum (Coleoptera: Tenebrionidae). J Mol Evol. 1999, 48: 615-624. 10.1007/PL00006504.
da Affonso RS, Affonso RS, Guimarães AP, Oliveira AA, Slana GBC, França TCC: Applications of molecular modeling in the design of new insect repellents targeting the odorant binding protein of Anopheles gambiae. J Braz Chem Soc. 2013, 24: 473-482. 10.1590/S0103-50532013000300015.
Ridley AW, Hereward JP, Daglish GJ, Raghu S, Collins PJ, Walter GH: The spatiotemporal dynamics of Tribolium castaneum (Herbst): adult flight and gene flow. Mol Ecol. 2011, 20: 1635-1646. 10.1111/j.1365-294X.2011.05049.x.
Langmead B, Salzberg SL: Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012, 9: 357-359. 10.1038/nmeth.1923.
Forêt S, Wanner KW, Maleszka R: Chemosensory proteins in the honey bee: Insights from the annotated genome, comparative analyses and expressional profiling. Insect Biochem Mol Biol. 2007, 37: 19-28. 10.1016/j.ibmb.2006.09.009.
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997, 25: 3389-3402. 10.1093/nar/25.17.3389.
Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JGR, Korf I, Lapp H, Lehväslaiho H, Matsalla C, Mungall CJ, Osborne BI, Pocock MR, Schattner P, Senger M, Stein LD, Stupka E, Wilkinson MD, Birney E: The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002, 12: 1611-1618. 10.1101/gr.361602.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R: The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009, 25: 2078-2079. 10.1093/bioinformatics/btp352.
Patel RK, Jain M: NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data. PLoS One. 2012, 7: e30619-10.1371/journal.pone.0030619.
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A: Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011, 29: 644-652. 10.1038/nbt.1883.
Stanke M, Diekhans M, Baertsch R, Haussler D: Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008, 24: 637-644. 10.1093/bioinformatics/btn013.
iBeetle consortium: http://ibeetle.uni-Goettingen.de,
Stanke M, Bucher G, Klingler M, iBeetle genome browser: http://bioinf.uni-Greifswald.de/tcas/,
Kent WJ: BLAT—The BLAST-Like Alignment Tool. Genome Res. 2002, 12: 656-664. 10.1101/gr.229202. Article published online before March 2002.
Petersen TN, Brunak S, von Heijne G, Nielsen H: SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011, 8: 785-786. 10.1038/nmeth.1701.
Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH: CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39 (Database issue): D225-D229.
Pavlidis P, Noble WS: Matrix2png: a utility for visualizing matrix data. Bioinformatics. 2003, 19: 295-296. 10.1093/bioinformatics/19.2.295.
Inkscape: http://Www.inkscape.org,
R: A Language and Environment for Statistical Computing. [http://www.R-project.org]
Anders S, Huber W: Differential expression analysis for sequence count data. Genome Biol. 2010, 11: R106-10.1186/gb-2010-11-10-r106.
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J: Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004, 5: R80-10.1186/gb-2004-5-10-r80.
Katoh K, Kuma K, Toh H, Miyata T: MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33: 511-518. 10.1093/nar/gki198.
Stamatakis A: RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinforma Oxf Engl. 2006, 22: 2688-2690. 10.1093/bioinformatics/btl446.
Kapushesky M, Adamusiak T, Burdett T, Culhane A, Farne A, Filippov A, Holloway E, Klebanov A, Kryvych N, Kurbatova N, Kurnosov P, Malone J, Melnichuk O, Petryszak R, Pultsin N, Rustici G, Tikhonov A, Travillian RS, Williams E, Zorin A, Parkinson H, Brazma A: Gene Expression Atlas update–a value-added database of microarray and sequencing-based functional genomics experiments. Nucleic Acids Res. 2011, 40: D1077-D1081.
Letunic I, Bork P: Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007, 23: 127-128. 10.1093/bioinformatics/btl529.
Wogulis M, Morgan T, Ishida Y, Leal WS, Wilson DK: The crystal structure of an odorant binding protein from Anopheles gambiae: Evidence for a common ligand release mechanism. Biochem Biophys Res Commun. 2006, 339: 157-164. 10.1016/j.bbrc.2005.10.191.
Zhou J-J, Robertson G, He X, Dufour S, Hooper AM, Pickett JA, Keep NH, Field LM: Characterisation of Bombyx mori Odorant-binding Proteins Reveals that a General Odorant-binding Protein Discriminates Between Sex Pheromone Components. J Mol Biol. 2009, 389: 529-545. 10.1016/j.jmb.2009.04.015.
Vieira FG, Forêt S, He X, Rozas J, Field LM, Zhou J-J: Unique Features of Odorant-Binding Proteins of the Parasitoid Wasp Nasonia vitripennis Revealed by Genome Annotation and Comparative Analyses. PLoS One. 2012, 7: e43034-10.1371/journal.pone.0043034.
Zhou J-J: Chapter Ten - Odorant-Binding Proteins in Insects. Vitam Horm. Volume Volume 83. Edited by: Litwack G. 2010, Amsterdam: Academic Press, 241-272. [Pheromones]
Eirín-López JM, Rebordinos L, Rooney AP, Rozas J: The Birth-and-Death Evolution of Multigene Families Revisited. Genome Dyn. Volume 7. Edited by: Garrido-Ramos MA. 2012, Basel: S. KARGER AG, 170-196.
Zhou J-J, Huang W, Zhang G-A, Pickett JA, Field LM: “Plus-C” odorant-binding protein genes in two Drosophila species and the malaria mosquito Anopheles gambiae. Gene. 2004, 327: 117-129. 10.1016/j.gene.2003.11.007.
Dreyer D, Vitt H, Dippel S, Goetz B, El Jundi B, Kollmann M, Huetteroth W, Schachtner J: 3D Standard Brain of the Red Flour Beetle Tribolium Castaneum: A Tool to Study Metamorphic Development and Adult Plasticity. Front Syst Neurosci. 2010, 4: 3-
Alabi T, Marion-Poll F, Danho M, Mazzucchelli GD, De Pauw E, Haubruge E, Francis F: Identification of taste receptors and proteomic characterization of the antenna and legs of Tribolium brevicornis, a stored food product pest. Insect Mol Biol. 2013, 23: 1-12.
Engsontia P, Sanderson AP, Cobb M, Walden KKO, Robertson HM, Brown S: The red flour beetle’s large nose: an expanded odorant receptor gene family in Tribolium castaneum. Insect Biochem Mol Biol. 2008, 38: 387-397. 10.1016/j.ibmb.2007.10.005.
Li J, Lehmann S, Weißbecker B, Ojeda Naharros I, Schütz S, Joop G, Wimmer EA: Odoriferous Defensive Stink Gland Transcriptome to Identify Novel Genes Necessary for Quinone Synthesis in the Red Flour Beetle, Tribolium castaneum. PLoS Genet. 2013, 9: e1003596-10.1371/journal.pgen.1003596.
Xu PX, Zwiebel LJ, Smith DP: Identification of a distinct family of genes encoding atypical odorant-binding proteins in the malaria vector mosquito, Anopheles gambiae. Insect Mol Biol. 2003, 12: 549-560. 10.1046/j.1365-2583.2003.00440.x.
Gong D-P, Zhang H-J, Zhao P, Xia Q-Y, Xiang Z-H: The Odorant Binding Protein Gene Family from the Genome of Silkworm, Bombyx mori. BMC Genomics. 2009, 10: 332-10.1186/1471-2164-10-332.
Vieira FG, Sánchez-Gracia A, Rozas J: Comparative genomic analysis of the odorant-binding protein family in 12 Drosophila genomes: purifying selection and birth-and-death evolution. Genome Biol. 2007, 8: R235-10.1186/gb-2007-8-11-r235.
Librado P, Rozas J: Uncovering the functional constraints underlying the genomic organisation of the Odorant-Binding Protein genes. Genome Biol Evol. 2013, 5: 2096-2108. 10.1093/gbe/evt158.
Jafari S, Alkhori L, Schleiffer A, Brochtrup A, Hummel T, Alenius M: Combinatorial Activation and Repression by Seven Transcription Factors Specify Drosophila Odorant Receptor Expression. PLoS Biol. 2012, 10: e1001280-10.1371/journal.pbio.1001280.
Leal WS, Nikonova L, Peng G: Disulfide structure of the pheromone binding protein from the silkworm moth, Bombyx mori. FEBS Lett. 1999, 464: 85-90. 10.1016/S0014-5793(99)01683-X.
Hungate EA, Earley EJ, Boussy IA, Turissini DA, Ting C-T, Moran JR, Wu M-L, Wu C-I, Jones CD: A Locus in Drosophila sechellia Affecting Tolerance of a Host Plant Toxin. Genetics. 2013, 195: 1063-1075. 10.1534/genetics.113.154773.
Qiao H-L, Deng P-Y, Li D-D, Chen M, Jiao Z-J, Liu Z-C, Zhang Y-Z, Kan Y-C: Expression analysis and binding experiments of chemosensory proteins indicate multiple roles in Bombyx mori. J Insect Physiol. 2013, 59: 667-675. 10.1016/j.jinsphys.2013.04.004.
Ju Q, Li X, Jiang X-J, Qu M-J, Guo X-Q, Han Z-J, Li F: Transcriptome and Tissue-Specific Expression Analysis of Obp and Csp Genes in the Dark Black Chafer. Arch Insect Biochem Physiol. 2014, 87: 177-200. 10.1002/arch.21188.
Qiao H, He X, Schymura D, Ban L, Field L, Dani FR, Michelucci E, Caputo B, della Torre A, Iatrou K, Zhou J-J, Krieger J, Pelosi P: Cooperative interactions between odorant-binding proteins of Anopheles gambiae. Cell Mol Life Sci. 2010, 68: 1799-1813.
Schultze A, Pregitzer P, Walter MF, Woods DF, Marinotti O, Breer H, Krieger J: The Co-Expression Pattern of Odorant Binding Proteins and Olfactory Receptors Identify Distinct Trichoid Sensilla on the Antenna of the Malaria Mosquito Anopheles gambiae. PLoS One. 2013, 8: e69412-10.1371/journal.pone.0069412.
Contreras E, Rausell C, Real MD: Proteome Response of Tribolium castaneum Larvae to Bacillus thuringiensis Toxin Producing Strains. PLoS One. 2013, 8: e55330-10.1371/journal.pone.0055330.
Xu J, Baulding J, Palli SR: Proteomics of Tribolium castaneum seminal fluid proteins: Identification of an angiotensin-converting enzyme as a key player in regulation of reproduction. J Proteomics. 2013, 78: 83-93.
Sirot LK, Poulson RL, McKenna MC, Girnary H, Wolfner MF, Harrington LC: Identity and transfer of male reproductive gland proteins of the dengue vector mosquito, Aedes aegypti: potential tools for control of female feeding and reproduction. Insect Biochem Mol Biol. 2008, 38: 176-189. 10.1016/j.ibmb.2007.10.007.
Ban L, Napolitano E, Serra A, Zhou X, Iovinella I, Pelosi P: Identification of pheromone-like compounds in male reproductive organs of the oriental locust Locusta migratoria. Biochem Biophys Res Commun. 2013, 437: 620-624. 10.1016/j.bbrc.2013.07.015.
Liu GX, Xuan N, Chu D, Xie HY, Fan ZX, Bi YP, Picimbon J-F, Qin YC, Zhong ST, Li YF, Gao ZL, Pan WL, Wang GY, Rajashekar B: Biotype expression and insecticide response of Bemisia tabaci chemosensory protein-1. Arch Insect Biochem Physiol. 2014, 85: 137-151. 10.1002/arch.21148.
Xuan N, Guo X, Xie H-Y, Lou Q-N, Lu X-B, Liu G-X, Picimbon J-F: Increased expression of CSP and CYP genes in adult silkworm females exposed to avermectins. Insect Sci. 2014, (doi: 10.1111/1744-7917.12116)
Liu Y-L, Guo H, Huang L-Q, Pelosi P, Wang C-Z: Unique function of a chemosensory protein in the proboscis of two Helicoverpa species. J Exp Biol. 2014, 217 (Pt 10): 1821-1826.
Findlay GD, Yi X, MacCoss MJ, Swanson WJ: Proteomics Reveals Novel Drosophila Seminal Fluid Proteins Transferred at Mating. PLoS Biol. 2008, 6: e178-10.1371/journal.pbio.0060178.
GEO accession. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63162,
endmemo: http://www.endmemo.com/bio/proie.php,
jpred: http://www.compbio.dundee.ac.uk/www-jpred/index.html,
Acknowledgements
We thank the transcriptome analysis laboratory (TAL, University Medical Center Göttingen, Germany), especially Gabriela Salinas-Riester for sequencing and technical support, Lotte Sogaard-Andersen from the Max Planck Institute for Terrestrial Microbiology (Marburg, Germany) for providing the MALDI TOF, and Montserrat Torres Oliva for technical and bioinformatics advice. This work was partially funded by the DFG Priority Programme “Integrative Analysis of Olfaction” (SPP 1392, SHA 678/13-1, SCHU 1135/13-1, and WI 1797/4-1).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
EAW, SS, JS, SA, and SD conceived and initiated the project. SD, SA, and JK performed the experiments. SD, GO, and LO analyzed the data and performed the bioinformatics. LG and MS provided the gene sets and browser presentation. SD created the figures and wrote the first draft of the manuscript. SD and EAW revised the manuscript. All authors critically read, commented on, and approved of the manuscript.
Stefan Dippel, Georg Oberhofer contributed equally to this work.
Electronic supplementary material
12864_2014_6882_MOESM1_ESM.xlsx
Additional file 1: Table S1: New names (column 1; uploaded at iBeetle genome browser [48]) based on chromosomal localization and corresponding previous names from Richards et al. 2008 [7] (column 2; uploaded at Beetle Base) as well as Foret and Maleska [16, 63] respectively from Vieira and Rozas [12] (column 3). The AA sequence including the signal peptide with the tryptic fragments identified by MALDI-TOF MS highlighted in red (column 4). The length of the pre-peptide (column 5) and mature peptide (column 6) in AA. The predicted length of the signal peptide (column 7; based on SignalP4.1 [50]). The molecular mass of the pre-peptide (column 8) and the mature peptide (column 9). The superfamily identified by the conserved domain database [51] (column 10), as well the probability as e-value (column 11). The confidence of the signal peptide [50] (column 12). The number of cysteines in the mature peptide (column 13). The Cysteine formula adjusted to the six highly conserved cysteines (column 14; including C-OBPs). The isoelectric point (based on endmemo [116], column 15). The position of alpha helices (based on jpred [117], column 16). Whether it was confirmed by cloning from cDNA (column 17). Primer sequences used for cloning from cDNA (columns 18 and 19). The reads per kilobase of exon model per million mapped reads of the different tissue samples (columns 20–34) The calculated fold-changes over body, and the corresponding p-values (DESeq package [80], column 35–42). (XLSX 74 KB)
12864_2014_6882_MOESM2_ESM.pdf
Additional file 2: Figure S1: Midpoint-rooted phylogenetic tree of CSP (A) and OBP (B) sequences from Tribolium castaneum (green branches), Drosophila melanogaster (red branches), and Anopheles gambiae (blue branches). Outer rings represent the expression in body, ‘mouthparts’ (Tribolium: palps, mandible, labrum and labium; Drosophila:palp and proboscis; Anopheles: maxillary palp) and antenna as percentage compared to the highest expressed gene according to the scale in the right middle. Please note that the methods used to obtain the different expression data (RNAseq and microarray) are not directly comparable. Thus, this figure can only give an impression of the tissue-specific abundance of the transcripts. The scale bars within the trees represent 1 amino acid substitution per site. Inner ring in B indicates the phylogenetic subclass (classic in black, ABPII in grey, C-OBP in green, C + OBP in blue, D7 in yellow, Dimer in red). Numbers on branches show values of 100 times replication bootstrap analysis higher than 70. (PDF 504 KB)
12864_2014_6882_MOESM3_ESM.xlsx
Additional file 3: Table S2: Proteins of antennal extracts identified by MALDI-TOF-TOF MS-fingerprinting: accession number, molecular weight, protein isoelectric point, peptide count, ion score, and Pfam prediction. (XLSX 24 KB)
12864_2014_6882_MOESM4_ESM.pdf
Additional file 4: Figure S2: Comparison of expression level of CSPs and OBPs in male and female heads (missing antennae but including mouthparts). Scatter plot of the RPKM values of the CSPs (in pink) and OBPs (classic in black, ABPII in grey, C-OBP in green, C + OBP in blue). The dotted lines represent a fivefold difference. (PDF 88 KB)
12864_2014_6882_MOESM5_ESM.pdf
Additional file 5: Figure S3: Comparison of expression level of CSPs and OBPs in male and female mouthparts. Scatter plot of the RPKM values of the CSPs (in pink) and OBPs (classic in black, ABPII in grey, C-OBP in green, C + OBP in blue). The dotted lines represent a fivefold difference. (PDF 87 KB)
12864_2014_6882_MOESM6_ESM.pdf
Additional file 6: Figure S4: Comparison of expression level of CSPs and OBPs in male and female legs. Scatter plot of the RPKM values of the CSPs (in pink) and OBPs (classic in black, ABPII in grey, C-OBP in green, C + OBP in blue). The dotted lines represent a fivefold difference. (PDF 84 KB)
12864_2014_6882_MOESM7_ESM.pdf
Additional file 7: Figure S5: Comparison of expression level of CSPs and OBPs in male and female bodies. Scatter plot of the RPKM values of the CSPs (in pink) and OBPs (classic in black, ABPII in grey, C-OBP in green, C + OBP in blue). The dotted lines represent a five fold difference. (PDF 84 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Dippel, S., Oberhofer, G., Kahnt, J. et al. Tissue-specific transcriptomics, chromosomal localization, and phylogeny of chemosensory and odorant binding proteins from the red flour beetle Tribolium castaneum reveal subgroup specificities for olfaction or more general functions. BMC Genomics 15, 1141 (2014). https://doi.org/10.1186/1471-2164-15-1141
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-15-1141