Abstract
Background
Clupeid fisheries in Lake Tanganyika (East Africa) provide food for millions of people in one of the world’s poorest regions. Due to climate change and overfishing, the clupeid stocks of Lake Tanganyika are declining. We investigate the population structure of the Lake Tanganyika sprat Stolothrissa tanganicae, using for the first time a genomic approach on this species. This is an important step towards knowing if the species should be managed separately or as a single stock. Population structure is important for fisheries management, yet understudied for many African freshwater species. We hypothesize that distinct stocks of S. tanganicae could be present due to the large size of the lake (isolation by distance), limnological variation (adaptive evolution), or past separation of the lake (historical subdivision). On the other hand, high mobility of the species and lack of obvious migration barriers might have resulted in a homogenous population.
Results
We performed a population genetic study on wild-caught S. tanganicae through a combination of mitochondrial genotyping (96 individuals) and RAD sequencing (83 individuals). Samples were collected at five locations along a north-south axis of Lake Tanganyika. The mtDNA data had low global FST and, visualised in a haplotype network, did not show phylogeographic structure. RAD sequencing yielded a panel of 3504 SNPs, with low genetic differentiation (FST = 0.0054; 95% CI: 0.0046–0.0066). PCoA, fineRADstructure and global FST suggest a near-panmictic population. Two distinct groups are apparent in these analyses (FST = 0.1338 95% CI: 0.1239,0.1445), which do not correspond to sampling locations. Autocorrelation analysis showed a slight increase in genetic difference with increasing distance. No outlier loci were detected in the RADseq data.
Conclusion
Our results show at most very weak geographical structuring of the stock and do not provide evidence for genetic adaptation to historical or environmental differences over a north-south axis. Based on these results, we advise to manage the stock as one population, integrating one management strategy over the four riparian countries. These results are a first comprehensive study on the population structure of these important fisheries target species, and can guide fisheries management.
Similar content being viewed by others
Introduction
Freshwater ecosystems support more species per unit area than any other ecosystem. Yet, they currently suffer from fast declines in species richness [1]. The decline in biodiversity reduces the resilience of aquatic ecosystems, decreasing their ability to provide ecosystem services such as food, drinking water, climate regulation, and social and health benefits [2]. As freshwater habitats play an important role in fisheries with almost 13% of the world’s aquatic catches [3], and one third of African fish catches [4], this decrease in resilience jeopardizes the future of human communities [5]. Therefore, it is unfortunate that freshwater fisheries have been less well studied compared to marine fisheries and are often overlooked in policy and regulation matters [6].
The sustainable exploitation of freshwater ecosystem services benefits from science-based management, based on sound biological knowledge of the system and its species. An important component of biological information is related to the structure of fish populations. The genetic structure of fish populations can be used to support the delineation of demographic units [7, 8], commonly referred to as stocks. Knowledge about stocks allows to preserve genetic variation and to decide on the size of meaningful management units [9]. Currently, most fisheries management units are not sufficiently supported by information on the population structure of the target species [10, 11]. Lack of scientifically supported management entails a risk for overfishing, and loss of population densities [12], especially when catch effort is not spread homogeneously [11].
In tropical systems, the biological knowledge on fisheries target species is less advanced and information on the population structure is often lacking. Hence, the scope for science-based management is small. This also holds for the Great Lakes of East Africa, in spite of their ecological, economic and social significance. Lake Tanganyika (LT) is the oldest African Great Lake, in which unique and very diverse aquatic communities have evolved [13]. It is situated in the western range of the Great African rift valley, measures almost 680 km in length and 50 km in width, and contains more than 1.89 × 107 km3 of water [14]. The oxygenated layer is deeper in the South (180 m) than in the North (120 m), as recorded during a dry season sampling [15]. The prevailing south-eastern winds cause an inclination of the thermocline, causing the upper water column to be somewhat warmer in the North (average annual temperature 25.8 °C), than in the South (average annual temperature 24 °C) [15, 16]. These differences are more pronounced in the dry season from May until September [16]. The lake is divided into three subbasins, which have been intermittently disconnected during periods of low water levels during its 6 million year history, forming distinct palaeolakes. The presumed prolonged division of the lake into these palaeolakes, approximately 1 million years ago, had profound influences on the lake’s diverse benthic fauna [17]. Lake levels continued to rise and fall, but it is assumed that since 106.000 years ago (106 kya), the subbasins of Lake Tanganyika have remained connected [18].
The fishery of LT plays an invaluable role in food security in one of the poorest regions in the world. Many people living near the lakeshore depend on artisanal fishing for their protein supply [19]. The Lake’s pelagic fisheries have a huge importance to local communities by providing almost 200,000 tons of fish yearly [20]. Pelagic catches are composed of mainly three species. The clupeids Stolothrissa tanganicae (Lake Tanganyika sprat; Clupeidae; Actinopterygii) and Limnothrissa miodon (Lake Tanganyika sardine; Clupeidae; Actinopterygii) provide 65% of the catch (by weight) [20]; a perciform predator, Lates stappersii (sleek lates; Latidae; Actinopterygii), provides 30% of the catch [20]. Additionally, S. tanganicae serves as an important food source for L. miodon and L. stappersii [21]. In the northern part of LT, S. tanganicae dominates the catches of artisanal fishermen [22]. In the South, the species is less abundant and catches are dominated by L. stappersii [23]. Stolothrissa tanganicae has a life style that is reminiscent of that of marine clupeids. It forms schools that differ in size and density throughout the day [24]. The species migrate deeper into the lake at dawn and back to the surface at dusk, probably following their zooplankton prey [21] and escaping their predators. The fish live up to 1.5 years, reach maturity at about 70 mm standard length (SL) [25] and their maximum SL is about 100 mm [23]. Stolothrissa tanganicae spawns throughout the year, with peaks in February–May [26] or August–September [27] in the North of the lake and in August–December [28], and possibly April–July [29] in the South. Eggs are spawned pelagically, sink and hatch one to 1.5 days later before they have reached the anoxic zone [24]. Feeding habits have mostly been studied in the northern part of the lake, where S. tanganicae feeds on zooplankton, mainly the calanoid copepod Tropodiaptomus simplex [30].
Observations at landing sites have shown a decrease of clupeid catches in LT [31, 32]. Hence, multiple calls for better management of this unique resource have been made [33, 34]. Yet, prior to this, it is necessary to understand the genetic structure of the two species, as it is unclear if they should be treated as single stocks or to be managed as different populations. A collapse of the clupeid fisheries would threaten the food security of millions of people. Additionally, loss of clupeid fisheries will also harm the biodiversity in LT as people will turn to fishing less resilient species, such as littoral cichlids. Furthermore, agriculture could increase further to compensate for the loss of protein source, which will cause runoff, destroying important habitats. Overall, the fisheries of LT are data-poor, which hampers the assessment of the exploitation status of the targeted stocks [35]. Clupeids can be considered very resistant to fisheries collapses because of their early age at maturity, pelagic lifestyle (reducing the risk of habitat destruction) and their absence in the bycatch of other species [36]. Nevertheless, there are many examples of pelagic species that were thought to be resilient against population collapses, yet collapsed under excessive fishing pressure. Among these examples are clupeids like the Pacific sardine (Sardinops sagax) [37, 38], and the Atlantic herring (Clupea harengus) [39].
Previous attempts to reveal the population genetic structure of S. tanganicae and L. miodon are scarce. In S. tanganicae, the only genetic study conducted so far suggested a single panmictic stock [40], while in L. miodon, no clear large-scale geographic structure could be identified [41]. However, the genetic markers used in these studies (RAPD markers in S. tanganicae; allozyme markers and mtDNA Restriction Fragment Length Polymorphism (RFLP) of the ND 5/6 gene in L. miodon), may lack the sensitivity to detect genetic structure in highly dispersive organisms. Recent developments in sequencing technologies, such as Restriction site Associated DNA markers (RAD sequencing) allow to infer population structure based on numerous single nucleotide polymorphisms (SNPs) [42]. The accuracy of RAD sequencing in detecting low levels of genetic differentiation therefore exceeds the accuracy of molecular techniques based on other marker types, even at small sample sizes [40, 43]. Although commonly used to detect population structure in pelagic marine species, RAD sequencing has less often been used in pelagic freshwater species.
In this study, we combine an analysis of mitochondrial DNA (mtDNA) haplotypes with a genomic analysis of nuclear DNA to assess population structure of S. tanganicae along a north-south axis in LT. A heterogeneous population genetic structure is possible for three reasons. First, the distance between the northern and southern end of the lake is large, compared to the assumed migration distances of this species, so levels of mixing might decrease with distance (hypothesis of isolation by distance). Second, there are limnological differences between the North and the South of the lake, to which S. tanganicae might have distinct adaptations (hypothesis of adaptive evolution). Since the eggs slowly sink to a depth of 150 m in the South [20], and need to hatch before reaching the anoxic zone, we assume this leaves less time for the eggs in the North to develop. The inclination of the thermocline could have an effect on larval development and productivity. Finally, fluctuations in the lake water level stands [41] have affected connectivity in the lake. Lower connectivity limits migration, which could lead to population structuring (hypothesis of historical subdivision). Alternatively, since migration distances are not exactly known and these sardines are highly mobile and may have large effective population size, the species could be panmictic across the entire north-south axis. This panmixia would fit with observations in other sardines and anchovies, which often show low population differentiation [44].
Material and methods
Sampling and DNA extraction
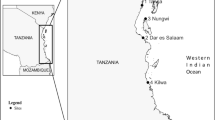
We selected five sampling sites along a latitudinal gradient covering the three subbasins of LT (Fig. 1). Two sites were selected in the northern basin (Uvira and Uvira 2), one in the central basin (Kalemie), and two in the southern basin (Mpulungu and Kalambo Lodge) (Table 1). This allowed us to evaluate population structure at the level of the entire lake, as well as among nearby sampling sites in the North and South of the lake. All samples were bought in the morning between August 11th and 20th 2016 (Table 1) from local fishermen who operated in a small range around the landing site. Since fishermen do not recast nets after they have been filled up by a passing school, all individuals within a sample belonged to the same school. To minimize the probability that a migrating school was sampled twice, the fish were bought on the same day for the two locations in the North, and on consecutive days for the two locations in the South (Table 1). For each sample, a finclip was stored in 99% ethanol. All individuals where measured and 32 were sexed of which 4 were male, 12 were female and 16 were not mature thus sex could not be identified. In total 96 individuals were used for the analysis of mitochondrial data and for the RAD library construction. DNA was extracted from finclips, using the NucleoSpin Tissue kit (Macherey-Nagel GmBH) according to the manufacturer’s instructions.
Map of Lake Tanganyika with sampling sites for Stolothrissa tanganicae. Uvira and Uvira2 are located in the northern subbasin, Kalemie in the central subbasin and Mpulungu and Kalambo Lodge are in the southern subbasin. Map made with Simple Mapper (http://research.amnh.org/pbi/maps/)
Mitochondrial sequence data
The mitochondrial cytochrome c oxidase subunit I (COI) gene was amplified using the universal primer combination HCO2198 (5’-TAAACTTCAGGGTGACCAAAAAATCA-3′) and LCO1490 (5’-GGTCAACAAATCATAAAGATATTGG-3′) [45]. The PCR mix consisted of 1 μL of template DNA, 2.5 μL PCR buffer, 0.75 μL Platinum MgCl2 (50 mM), 0.5 μL of dNTPs (10 mM), 1 μL of both primers (10 μM), 0.15 μL Platinum Taq polymerase (5 units/μL) and 18.1 μL of milli-Q water, totaling 25 μL. The PCR cycling profile consisted of 3 min at 94 °C, followed by 35 cycles at 94 °C for 45 s, 52 °C for 40 s, 72 °C for 90 s, 10 min at 72 °C and cooling to 4 °C. PCR products were purified by means of GFX purification columns (GE Healthcare, Chicago, IL, USA), subjected to sequencing reactions using the BigDye v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) and sequenced using the LCO1490 primer, with an ABI Prism 3100 Genetic Analyzer (Applied Biosystems). Sequence quality was verified with Geneious v11 [46] and MEGA v7.0 [47] by checking each SNP for base quality, assuming a reading error if a SNP is rare and quality is low. We checked for mutations recorded on the second position in a codon, which did not occur. Sequences were aligned with MUSCLE [48] using the default settings (Gap penalties: open = − 400; extend = 0, clustering method UPGMB, λ = 24). Before analyses, primers were trimmed out and sequences translated into amino acids to check for the absence of internal stop codons. Given the absence of gaps, the alignment was straightforward. The mitochondrial sequence data were used (a) to double-check the morphological identification of voucher specimens via DNA barcoding (data not shown) and (b) to assess possible genetic structure across individuals from different sampling sites. For this, a Median Joining Network [49] was made with PopART 1.7 [50], with ε = 0. Differentiation among individuals from the different sampling sites was estimated by global FST and pairwise FST between sampling sites in the diveRsity package [51] in R, using 100 bootstraps to calculate bias corrected 95% confidence intervals. We calculated number of haplotypes and Tajima’s D statistic, using DnaSP v6 [52].
RAD library preparation
Six RAD libraries, each including 16 individually indexed specimens, were prepared according to the protocol described in Baird et al. [53] and Etter et al. [54]. Individual DNA samples were digested using restriction enzyme SbfI-HF (NEB, cut site 5’-CCTGCA^GG-3′). In silico digestion of the genome of the related Atlantic herring (Clupea harengus) [55] revealed 21,544 RAD loci with SbfI. Samples were individually barcoded with P1 adapters ligated to the fragment’s overhanging end. The RAD libraries were sheared to a size of 350 base pairs (bp) and the fragments between 200 and 700 bp selected by gel size selection. A second, library-specific barcoded adapter (P2), was ligated to the DNA fragments for identification of the samples. RAD libraries were sequenced 101 bp paired-end on an Illumina HiSeq1500 platform at the Medical Centre for Genetics of the University of Antwerp, Belgium.
Processing of RAD data
Overall read quality was assessed using the FastQC software v0.11.5 [56]. Raw sequence data was demultiplexed using the process_radtags module in Stacks v1.46 [57, 58], while reads characterized by ambiguous barcodes, ambiguous cut sites or low quality scores were discarded. PCR duplicates were removed via the clone_filter module and SNPs were called using the denovo_map pipeline, both implemented in Stacks. We screened a range of parameter combinations and selected a minimum coverage of ten reads per stack (m = 10) and a maximum number of five base pair differences between stacks within (M = 5) and between (n = 5) individuals. This parameter setting allowed us to retain a sufficient number of orthologues at a considerable depth. Individuals with insufficient raw reads (< 0.8 million), a high proportion of missing data (> 50%) and low depth (< 9.7) were removed. A final round of filtering was performed using VCFtools v0.1.14 [59] in order to discard sites characterized by heterozygosity excess (p-value < 0.01), a minimum allele frequency of less than 0.05, and more than 20% of missing data.
Neutral population structure
Genetic variation of each sample was assessed by expected and observed heterozygosity and allelic richness, using the diveRsity v1.9.90 package in R v3.4.1. Using the same package, estimates of global FST were calculated, for two geographic scales, lake-wide and between nearby locations. To make lake-wide comparisons, we pooled the two northern (Uvira + Uvira2) and the two southern (Mpulungu + Kalambo Lodge) sampling sites. Pairwise FST was calculated across the sampling sites, with 95% confidence intervals based on 100 bootstrap iterations over loci.
Population structure was inspected with the R package ADEGENET v2.1.0 [60] to perform a non-centered, non-scaled Principal Coordinates Analysis (PCoA) based on Euclidean distances between specimens. Missing data in this analysis were replaced by the mean allele frequencies. In addition, we performed a Discriminant Analysis of Principal Components (DAPC) [61] with default settings. DAPC reduces the variation within the sampling sites, while maximizing the variation between them. As the amount in explained variance showed a continuous gradual decline with most important PCs, no optimal cutoff of number of PCs could be identified. Therefore, the DAPC was based on 28 PCs, the largest number of informative PCs [60].
Population structure was assessed using an MCMC method to infer recent shared ancestry based on patterns of genomic similarity implemented in fineRADstructure [62], which is a modification of fineSTRUCTURE [63] for RAD data. As this analysis showed to be highly sensitive to missing data, we retained only SNPs scored in more than 90% of all individuals. The RAD tags were ordered according to linkage disequilibrium with the sampleLD.R script provided in fineRADstructre. Subsequently, the co-ancestry matrix was calculated and used to identify populations by a clustering algorithm. This approach is robust for missing RAD alleles and is sensitive to subtle population structure. The MCMC chain ran with a burnin of 100,000, 100,000 iterations and a thinning interval of 1000. We further explored whether genetic similarity between individuals decreased with geographical distance by conducting a spatial autocorrelation analysis over the five sampling sites in GenAlEx v6.501 [64,65,66]. Correlation coefficients between individuals were depicted as a function of increasing inter-individual geographical distance and confidence intervals based on 1000 bootstraps.
Genome scan of outlier loci
Putative signatures of natural selection were assessed using three different approaches to detect outlier loci. First, we assessed the distribution of the global FST values among loci, at the lake-wide scale and between all five of the locations, to identify possible candidates for outliers, using the diveRsity v1.9.90 package of R. Secondly, we performed a Bayesian outlier detection method in BayeScan v2.1 [67] which incorporates locus- and population-specific FST effects [67,68,69]. For each level, three replicate runs were executed with default parameter settings. False discovery rate (FDR) threshold was 0.05, and only loci consistently identified as outliers in each of three independent runs were considered as true outliers. Finally, we assessed the possible occurrence of adaptation along the latitudinal gradient. We applied an individual-based latent fixed mixed model (LFMM) in which SNP frequencies were associated to latitudinal variation, while accounting for neutral population structure [70]. The number of latent factors was set to one. We ran the model ten times, using 20,000 sweeps for burn-in and 40,000 additional sweeps as run-length and calculated the median Z-value of all replicated runs for each locus separately. We applied a correction by dividing the raw p-value by a genomic inflation factor, corresponding to the median of the square z-value divided by the median of the chi-square distribution [71]. To correct for the multiple tests, SNPs that were considered as non-neutral, were characterized by a q-value of 0.05 or less [71]. LFMM analyses were performed using the LEA package in R [72].
Results
Phylogeography based on mitochondrial sequence data
The Median Joining Network based on mitochondrial COI fragments of a length of 643 bp, does not suggest separation either between the five sampling sites, or between the three subbasins (Fig. 2). The global FST value between sampling sites is 0.0026. Pairwise FST values (Table 2) do not significantly differ from zero. Values range from -0.027 (95% CI: - 0.072, 0.0641) (Mpulungu – Uvira 2) to 0.0327 (95% CI: -0.0358, 0.1498) (Kalemie – Uvira). In these 96 samples, there are 47 different haplotypes and Tajima’s D is significantly negative (D = -2.414, p < 0.01).
Haplotype network of COI sequences of Stolothrissa tanganicae (n = 96). Median Joining Network (ε = 0) created in PopART v1.7. Each circle represents a haplotype, the size of circles corresponds to the number of individuals with the haplotype. Colors indicate sampling sites. Bars indicate the number of mutations between two haplotypes. Small black circles indicate hypothetical haplotypes, predicted by the model. Uvira and Uvira 2 are in the northern basin, Kalemie is in the central basin and Mpulungu and Kalambo Lodge are in the southern basin
Quality of RAD genotyping
Due to a low number of reads (< 0.8 million), a high percentage of missing reads (> 50%) and low depth (< 9.7), 12 individuals were discarded. Two individuals from the Mpulungu sampling site were very similar, indicating possible contamination. To solve this, one of these individuals was removed. This resulted in 83 retained individuals, at least 15 per sampling site, with the number of reads per specimen ranging from 0.9 to 3.7 million (average per specimen = 1.96 million). Filtering produced a final dataset containing 3504 SNPs distributed across these 83 individuals, with a mean depth per individual of 29.66 (minimum of 9.7 and maximum of 57.9) and a mean missing per individual of 12% (minimum of 0.007% and maximum of 48%). Detailed information on missing data per individual can be found in Additional file 1.
Nuclear genetic diversity and neutral population structure
Observed and expected heterozygosity values were similar among the sampling sites, with expected heterozygosity ranging from 0.2088 (Kalemie) to 0.2605 (Uvira) and observed heterozygosity ranging from 0.1920 (Kalemie) to 0.2619 (Uvira) (Table 3). Allelic richness across the different sampling sites ranged from 1.7831 (Kalemie) to 1.9047 (Mpulungu) (Table 3).
Genetic differentiation estimated by global FST was relatively low, but significantly different from zero, with a FST value of 0.0068 (95% CI: 0.0057–0.0079) between sampling sites and a FST value of 0.0054 (95% CI: 0.0046–0.0066) between the northern, central and southern basin. Similarly, pairwise FST values between sampling sites are low, ranging from -0.0012 (95% CI: -0.002–0.0001) (Kalambo Lodge – Uvira) to 0.0250 (95% CI: 0.0215–0.0281) (Kalemie – Uvira) (Table 2).
The PCoA revealed no clustering based on the geographic origin of the samples (Fig. 3a). Individuals are separated on PC1 (9.30% explained variation) in one large cluster of 63 individuals and one smaller cluster of 20 individuals, regardless of the sampling site. PC2 and PC3 explained 1.72 and 1.71% of the variation respectively. PCoA was repeated with only the individuals in the larger cluster, to check if there is no hidden structure (Fig. 3b). Here, PC1 explains 2.47% of the variation and PC2 explains 2.44%. FST between the large and smaller cluster is significantly different from zero: 0.1338 [0.1239,0.1445]. The DAPC analysis shows no obvious pattern of genetic structuring across sampling sites, although some degree of separation on the diagonal is visible, with the samples from the central basin placed between those from the North and those from the South (Fig. 4, Additional file 2). For the visualization of patterns of haplotype similarity with fineRADstructure (Fig. 5), a reduced dataset of 1255 SNPs was used, to correct for effects of unevenly distributed levels of missing data. The structure provided by the fineRADstructure analysis corroborated with the results of the PCoA analysis, placing the same individuals in the same two clusters. These two groups are irrespective of sex or sampling site.
PCoA based on kinship of nuclear DNA. Each dot represents one S. tanganicae individual. Dots that are closer together have more similar genotypes. Colors represent the five sampling sites. a. all individuals, PC1 explains 9.30% of the variation and PC2 1.72% of the variation. b. Plot with only the individuals from the larger cluster, PC1 2.47% explains of the variation and PC2 explains 2.44% of the variation. Made with ADEGENET v2.1.0 package in R
FineRADstructure analysis for visualization of patterns of haplotype similarity: co-ancestry matrix based on a reduced dataset of 1255 SNPs. Colors indicate scale of relatedness between individuals, with yellow being low relatedness and blue/black indicating high relatedness. No structuring per sampling site is visible. A cluster of individuals is apparent in the upper right of the graph. These individuals correspond to the individuals that score high on the first axis of the PCoA plot, and are spread over the different sampling sites. Made with the fineRADstructure software [62]
Autocorrelation analysis shows a low but significant level of genetic structuring in the five sampling sites along a north-south axis of LT, indicating a difference in populations in the North compared to the South. At a distance of 400 km, random processes like stochastic drift seem to overcome the homogenizing effect of gene flow (Fig. 6).
Autocorrelation (r) showing genetic similarity over geographical distance. Error bars bound the 95% confidence interval as determined by bootstrap resampling. Over a distance of 400 km, 95% CI include zero, showing that random processes like stochastic drift overcome the homogenizing effect of gene flow. Analysis done in GenAlEx v6.501 [66]
Outlier loci
Patterns in global FST at each SNP are concordant with previous results as the majority of FST values are clustered around zero, indicating low levels of genetic structuring (Additional file 3). Only 32 SNPs are characterized with a FST higher than 0.1 according to sampling site and 12 SNPs according to subbasin. The highest FST value is 0.21. None of these are identified as significant outliers by BayeScan (Additional file 4) or LFMM (Additional file 5) at a FDR threshold of 0.05.
Discussion
Population structure of Stolothrissa tanganicae
The population structure of Stolothrissa tanganicae was explored over five sampling sites in the three subbasins of LT using mitochondrial COI sequences and RAD sequencing data, to verify the existence of biologically meaningful management units. This species showed for both marker types a very weak genetic structure, suggesting a near-panmictic population. For both markers, the difference between samples from the different subbasins is not larger than the difference within subbasins. This pattern is obvious in both the PCoA and fineRADstructure analysis of the RAD data and the haplotype network based on the mitochondrial DNA. The PCoA plot (Fig. 3) and the Median Joining Network (Fig. 2), show no genetic structuring according to sampling site or subbasin. Autocorrelation analysis revealed that there is a limitation to long-distance migration, as at a distance of 400 km, a decline in gene flow becomes apparent. The high number of different mitochondrial haplotypes (47), suggests many different maternal lineages.
The diverse set of lineages and the overall weak genetic structure confirm the conclusions of a previous population genetic study on S. tanganicae, which suggested a single panmictic stock [40]. However, this study was based on random amplified polymorphic DNA (RAPD) markers. RAPD markers are often difficult to interpret, and the results are not always reproducible. Our confirmation of the results based on a large set of high-quality SNPs represents an important benchmark, and indicates that S. tanganicae has been near-panmictic since the 1990s. No other population genetic studies on S. tanganicae are available, but a study by Sako et al. [43] revealed significant differences in otolith chemistry between populations from the northern and southern basin. This difference suggests that populations from the North and South of the lake spend most of their lifetime in different environments and implies that long-distance migrations must be rare. This seems to contradict the genetic patterns. However, a few migrants per generation are usually sufficient to maintain a near-panmictic population at the level of the entire lake.
Some of our analyses suggested the existence of two separate groups, independent of geographical origin. This is apparent in the PCoA plot (Fig. 3a), where we found a separation along the first axis. FineRADstructure analysis revealed the same two clusters. It is unclear what difference there is between these two groups, which differ in size. Missing data were equally distributed among the groups, so this is not the origin of the separation. The two groups could point to the two different sexes, yet for the 16 individuals that have been sexed in this study, male and female individuals show up in both groups in both analysis. Separate groups may as well arise because of a difference in spawning times. There are currently no indications for this in S. tanganicae, but it has been shown for Atlantic herring where spring-spawning and autumn spawning individuals were genetically differentiated [73]. Another possibility is that S. tanganicae frequently hybridizes with the other endemic clupeid, L. miodon, since FST between both groups is very large (FST = 0.1338 [0.1239,0.1445]). A larger sample size, as well as individuals of both species, are required to test the identity of the two separate groups. Individuals from the two groups have been found in all the sampling sites, so they do not alter the conclusions of the various analyses in this study.
The lakescape of pelagic fish
It is worthwhile to speculate what may cause the weak geographical population genetic structure of Stolothrissa tanganicae. At the start of this study, we hypothesized that population structure could arise due to isolation by distance, adaptive evolution, or the distinct history of the subbasins. We also considered the possibility of a homogeneous population because of large effective population sizes and high mobility of the species and the long period during which obvious migration barriers were absent. Our data did not show genetic differentiation between the different sampling locations over a north-south axis of the lake.
First, the data revealed a very weak pattern of isolation by distance, which was detected with the autocorrelation analyses. In 1970, Coulter stated, based on his observations in the northern and southern basins, that there were no reports of large clupeid migrations, and that there was no reason to assume there were any [25]. Yet, as stated above, some migration either individually or in schools may cause sufficient gene flow to keep the population structure near-panmictic. Similar to the marine environment, the pelagic zone of LT does not contain many barriers for migration. The frequent algal blooms in LT attract zooplankton, which in turn attracts the sprats. These algal blooms occur in the South of the lake in May–June, due to upwelling of nutrient rich water caused by tilting of the epilimnion because of strong south-east winds. After the winds cease around September, currents reverse and an algal bloom occurs in the North in October–November [16]. Migrations follow these blooms, as indicated by a positive correlation between S. tanganicae abundance and measures of chlorophyll a [23]. Catch statistics indicate a peak in S. tanganicae catches during phytoplankton blooms in the North [23, 74] and the South [75] of the lake. These seasonal migrations may contribute to the mixing of populations.
We found no traces of local adaptation to different conditions in the North and the South of LT. The number of loci in this study may have been too low to detect genomic regions involved in adaptive processes. There are some limnological differences along a north-south axis that could trigger local adaptation. For instance, the timing of major spawning events in S. tanganicae differ across the lake [21, 25, 28], but it is unknown if this difference in spawning time is an adaptive trait or linked to phenotypic plasticity in response to the timing of the plankton blooms [28] and depth of the oxygenated layer. Little is known about spawning areas and mating behaviour of the sprat. There is little information on how the eggs are fertilized and deposited and about dispersal of eggs, both possible facilitators of population mixing, as has been shown for marine species [76]. Expanding this limited knowledge is needed for good monitoring and conservation of the stock, and could help in explaining why the population remains homogeneous.
Our results do not show signatures of a population that differentiated because of historical barriers, which would have caused greater differences in genotypes between our samples. At times of extreme low-stands of the water levels of LT, the lake would be divided into three separate lakes, according to the three subbasins. It is assumed that the differentiation in cichlids was triggered by this isolation [77,78,79]. It is unclear if S. tanganicae also differentiated into different populations in the isolated subbasins as a result of low water levels. This lack of observed differentiation could be due to the pelagic life style of the sprat, enabling dispersal throughout the lake, similar to the benthopelagic Lake Tanganyika’s giant cichlid (Boulengerochromis microlepis) [80] and two eupelagic Bathybates species (B. fasciatus and B. leo) [81] whose populations also do not show any phylogeographic structure.
A possible explanation for the homogeneous structure found here for S. tanganicae is that these populations could have passed through a bottleneck and quickly expanded again. This assumption is supported by a significant negative value of Tajima’s D statistic, showing that observed heterozygosity is lower than expected heterozygosity due to inbreeding. Clupeids are known to have highly fluctuating population sizes, with large declines in numbers and fast expansions [82], leading to traceable bottlenecks [44]. Fishing pressure, poor recruitment or limited food availability could have significantly reduced the number of remaining sprats. Lake Tanganyika sprat is an r-selected species [83] with a short lifespan, many offspring and reaching an age of maturity within a few months [84]. Furthermore, schooling reduces the effort to find a mate. This makes S. tanganicae excellently equipped for rapid population expansions [20].
Just like S. tanganicae in this study, sardines worldwide, often assessed over greater geographic distances, show below-average levels of population differentiation in comparison to other marine fishes. This is generally explained by their pelagic lifestyle, limited proportion of the population that contributes to the next generation, overharvesting and population bottlenecks [44, 85, 86]. In some cases, population genetic structure was detected [87], for example in the presence of physical barriers such as ocean currents [88] or over large geographical distances [89]. In other cases, subtle levels of ecological adaptation have been detected, for example between Atlantic herring (Clupea harengus) from the North Sea [90] and the Baltic Sea [91].
Implications for fisheries management and future research
The weak genetic structure in S. tanganicae over a north-south axis of LT, emphasises the need for integrated management of the entire stock. On the one hand, a single homogeneous stock might be easier to manage, since local extinctions can be countered by migrations from other populations. The adaptive potential and chance of survival of a metapopulation is bigger than that of an isolated subpopulation. On the other hand, managing such a homogeneous population has its own difficulties: Lake Tanganyika is bordered by four countries, each with its own legislation, law enforcement and economic reality. As the geographically unstructured sprat stocks do not correspond to international borders, each local management regime influences the stock available to the neighbouring countries. Our findings also underpin the importance of locating and protecting the spawning areas of S. tanganicae, since degradation of a spawning area could impact the stock in a wider area. Illegal fishing of clupeid fry in the spawning areas forms a huge burden on the stocks [92, 93]. It is also important to have more knowledge on which parts of the lake serve as sources and which as sinks for the S. tanganicae population. This information is vital to delineate spawning areas and source populations as protected areas.
Future research on the pelagic species in Lake Tanganyika remains necessary to provide information for management and conservation. More information on migrations of these pelagic clupeids would be beneficial for more directed management. The availability of a reference genome would be a step towards interpretation of adaptive traits if outlier SNPs would be detected. It will also be vital towards discovering genomic signatures of overfishing. There is also a need to look at the population structure of the two other major fisheries target species in Lake Tanganyika, L. miodon and L. stappersii. They both have a more littoral lifestyle than S. tanganicae [21], hence their populations might be more structured. Also, L. stappersii has a very different life history than the clupeids: these predators are bigger and live longer, which might affect their population structuring. This type of research can be useful in many other systems. It can be expanded to pelagic fish of the other African Great Lakes, and beyond. There are many lake ecosystems where a small, fast growing, pelagic fish species forms the link between zooplankton and piscivorous animals, just like the clupeids of Lake Tanganyika. Many of these systems would benefit from having information about the population structure of their pelagic fisheries targets. In some of these lakes, for example Lake Victoria, the pelagic fishes are becoming more important in the ecosystem due to overfishing of the larger fish species.
Conclusion
Our study confirms previous findings on the population structure of S. tanganicae in Lake Tanganyika. A near-panmictic population structure was detected over a north-south axis of the lake, with slightly increasing genetic distance over increasing geographical distance. This homogeneity in the stock of one of the major fisheries target species in LT underscores the need for integrated stock management between the four nations bordering Lake Tanganyika.
Abbreviations
- bp:
-
Base pairs
- CI:
-
Confidence interval
- COI:
-
Cytochrome c oxidase subunit I
- FDR:
-
False discovery rate
- Kya:
-
Thousand years ago
- LFMM:
-
Latent fixed mixed model
- LT:
-
Lake Tanganyika
- mtDNA:
-
Mitochondrial DNA
- NGS:
-
Next Generation Sequencing
- RAD:
-
Restriction site associated DNA
- RFLP:
-
Restriction fragment length polymorphism
- SE:
-
Standard error
- SL:
-
Standard length
- SNP:
-
Single nucleotide polymorphism
References
Kopf RK, Finlayson CM, Humphries P, Sims NC, Hladyz S. Anthropocene baselines: assessing change and managing biodiversity in human-dominated aquatic ecosystems. Bioscience. 2015;65:798–811.
Chapin FS III, Zavaleta ES, Eviner VT, Naylor RL, Vitousek PM, Reynolds HL, et al. Consequences of changing biodiversity. Nature. 2000;405:234–42.
FAO. The State of World Fisheries and Aquaculture; 2016. p. 2016.
de Graaf GJ, Garibaldi L. The value of African fisheries. FAO. Fish. Aquac. In: Circ; 2014.
Ripple WJ, Wolf C, Galetti M, Newsome TM, Alamgir M, Crist E, et al. World scientists’ warning to humanity: a second notice. Bioscience. 2017;67:1026–8.
Cooke SJ, Allison EH, Beard TD, Arlinghaus R, Arthington AH, Bartley DM, et al. On the sustainability of inland fisheries: finding a future for the forgotten. Ambio. Springer Netherlands. 2016;45:753–64.
Stephenson RL. Stock complexity in fisheries management: a perspective of emerging issues related to population sub-units. Fish Res. 1999;43:247–9.
Hauser L, Carvalho GR. Paradigm shifts in marine fisheries genetics: ugly hypotheses slain by beautiful facts. Fish Fish. 2008;9:333–62.
Ovenden JR, Berry O, Welch DJ, Buckworth RC, Dichmont CM. Ocean’s eleven: a critical evaluation of the role of population, evolutionary and molecular genetics in the management of wild fisheries. Fish Fish. 2015;16:125–59.
Reiss H, Hoarau G, Dickey-Collas M, Wolff WJ. Genetic population structure of marine fish: mismatch between biological and fisheries management units. Fish Fish. 2009;10:361–95.
Guan W, Cao J, Chen Y, Cieri M, Quinn T. Impacts of population and fishery spatial structures on fishery stock assessment. Can J Fish Aquat Sci. 2013;70:1178–89.
Kerr LA, Hintzen NT, Cadrin SX, Clausen LW, Dickey-collas M, Goethel DR, et al. Lessons learned from practical approaches to reconcile mismatches between biological population structure and stock units of marine fish. ICES J Mar Sci. 2016;74:1708–22.
Salzburger W, Van Bocxlaer B, Cohen AS. Ecology and evolution of the African Great Lakes and their faunas. Annu Rev Ecol Evol Syst. 2014;45:519–45.
Huttula T. Flow, thermal regime and sediment transport studies in Lake Tanganyika. Huttula T, editor. Kuopio University Publications C. Natural and Environ Sci 73; 1997.
De Wever A, Muylaert K, Van Der Gucht K, Pirlot S, Cocquyt C, Descy J, et al. Bacterial community composition in Lake Tanganyika: vertical and horizontal heterogeneity. Appl Environ Microbiol. 2005;71:5029–37.
Plisnier PD, Chitamwebwa D, Mwape L, Tshibangu K, Langenberg V, Coenen E. Limnological annual cycle inferred from physical-chemical fluctuations at three stations of Lake Tanganyika. Hydrobiologia. 1999;407:45–58.
Danley PD, Husemann M, Ding B, DiPietro LM, Beverly EJ, Peppe DJ. The impact of the geologic history and paleoclimate on the diversification of east African cichlids. Int J Evol Biol. 2012;2012:1–20.
McGlue MM, Lezzar KE, Cohen AS, Russell JM, Tiercelin JJ, Felton AA, et al. Seismic records of late Pleistocene aridity in Lake Tanganyika, tropical East Africa. J Paleolimnol. 2008;40:635–53.
Van der Knaap M, Katonda KI, De Graaf GJ. Lake Tanganyika fisheries frame survey analysis: assessment of the options for management of the fisheries of Lake Tanganyika. Aquat Ecosyst Heal Manag. 2014;17:4–13.
Mölsä H, Reynolds JE, Coenen EJ, Lindqvist OV. Fisheries research towards resource management on Lake Tanganyika. Hydrobiologia. 1999;407:1–24.
Coulter GW. Lake Tanganyika and its life. In: British museum (natural history); 1991.
Kimirei IA, Mgaya YD. Influence of environmental factors on seasonal changes in clupeid catches in the Kigoma area of Lake Tanganyika. African J Aquat Sci. 2007;32:291–8.
Plisnier PD, Mgana H, Kimirei I, Chande A, Makasa L, Chimanga J, et al. Limnological variability and pelagic fish abundance (Stolothrissa tanganicae and Lates stappersii) in Lake Tanganyika. Hydrobiologia. 2009;625:117–34.
Matthes H. Preliminary investigations into the biology of the Lake Tanganyika Clupeidae. Fish Res Bull. 1967;4:1965–6.
Coulter G. Population changes within a group of fish species following their exploitation. J Fish Biol. 1970;2:329–53.
Roest FC. Stolothrissa tanganicae: population dynamics, biomass evolution and life history in the Burundi waters of Lake Tanganyika. UN, FAO, CIFA Tech Pap. 1977:42–63.
Mulimbwa N, Sarvala J, Raeymaekers JAM. Reproductive activities of two zooplanktivorous clupeid fish in relation to the seasonal abundance of copepod prey in the northern end of Lake Tanganyika. Belgian J Zool. 2014;144:77–92.
Chapman DW. Van well P. growth and mortality of Stolothrissa tanganicae. Trans Am Fish Soc. 1978;107:523–7.
Ellis CMA. The size at maturity and breeding seasons of sardines in southern Lake Tanganyika. AfrJTropHydrobiolFish. 1971;1:59–66.
Mgana HF, Herzig A, Mgaya YD. Diel vertical distribution and life history characteristics of Tropodiaptomus simplex and its importance in the diet of Stolothrissa tanganicae, Kigoma, Tanzania. Aquat Ecosyst Health Manag. 2014;17:14–24.
Coenen EJ, Nikomeze E. Lake Tanganyika, Burundi, results of the 1992–93 catch assessment surveys; 1994.
Van Der Knaap M, Kamitenga DM, Many LN, Tambwe AE, De Graaf GJ. Lake Tanganyika fisheries in post-conflict Democratic Republic of Congo. Aquat Ecosyst Health Manag. 2014;17:34–40.
Fryer G. Conservation of the Great Lakes of East Africa: a lesson and a warning. Biol Conserv. 1972;4:256–62.
Britton AW, Day JJ, Doble CJ, Ngatunga BP, Kemp KM, Carbone C, et al. Terrestrial-focused protected areas are effective for conservation of freshwater fish diversity in Lake Tanganyika. Biol Conserv. 2017;212:120–9.
Vasconcellos M, Cochrane K. Overview of world status of data-limited fisheries : inferences from landings statistics. In: Kruse GH, Gallucci VF, Hay DE, Perry RI, Peterman RM, Shirley TC, et al., editors. Fish assess Manag data-limited situations. Anchorage: Alaska Sea Grant College Program University of Alaska Fairbanks Lowell; 2005. p. 1–20.
Hutchings JA, Reynolds JD. Marine fish population collapses: consequences for recovery and extinction risk. Bioscience. 2004;54:297-309.
Murphy GI. Vital statistics of the Pacific sardine (Sardinops Caerulea) and the population consequences. Ecology. 1967;48:731–6.
Zwolinski JP, Emmett RL, Demer DA. Predicting habitat to optimize sampling of Pacific sardine (Sardinops sagax). ICES J Mar Sci. 2011;68:867–79.
Overholtz WJ. The Gulf of Maine - Georges Bank Atlantic herring (Clupea harengus): spatial pattern analysis of the collapse and recovery of a large marine fish complex. Fish Res. 2002;57:237–54.
Hess JE, Matala AP, Narum SR. Comparison of SNPs and microsatellites for fine-scale application of genetic stock identification of Chinook salmon in the Columbia River basin. Mol Ecol Resour. 2011;11:137–49.
Cohen AS, Talbot MR, Awramik SM, Dettman DL, Abell P. Lake level and paleoenvironmental history of Lake Tanganyika, Africa, as inferred from late Holocene and modern stromatolites. Bull Geol Soc Am. 1997;109:444–60.
Mesnick SL, Taylor BL, Archer FI, Martien KK, Treviño SE, Hancock-Hanser BL, et al. Sperm whale population structure in the eastern and central North Pacific inferred by the use of single-nucleotide polymorphisms, microsatellites and mitochondrial DNA. Mol Ecol Resour. 2011;11:278–98.
Puckett EE, Eggert LS. Comparison of SNP and microsatellite genotyping panels for spatial assignment of individuals to natal range: a case study using the American black bear (Ursus americanus). Biol Conserv. 2016;193:86–93.
Grant WS, Bowen BW. Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for coservation. J Hered. 1998;89:415–26.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–9.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7.
Bandelt H-J, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol. 1999;16:37–48.
Leigh JW, Bryant D. POPART: full-feature software for haplotype network construction. Methods Ecol Evol. 2015;6:1110–6.
Keenan K, Mcginnity P, Cross TF, Crozier WW, Prodöhl PA. diveRsity: an R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol Evol. 2013;4:782–8.
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017;34:3299–302.
Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 2008;3:1–7.
Etter PD, Bassham S, Hohenlohe PA, Johnson EA, Cresko WA. SNP discovery and genotyping for evolutionary genetics using RAD sequencing. Methods Mol Biol. 2011;772:157–78.
Barrio AM, Lamichhaney S, Fan G, Rafati N, Pettersson M, Zhang H, et al. The genetic basis for ecological adaptation of the Atlantic herring revealed by genome sequencing. elife. 2016;5:1–32.
Andrews S. FastQC: a quality control tool for high throughput sequence data; 2010.
Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH, De Koning D-J. Stacks: building and genotyping loci de novo from short-read sequences. G3journall. 2011;1:171–82.
Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. Stacks: an analysis tool set for population genomics. Mol Ecol. 2013;22:3124–40.
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–8.
Jombart T. Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–5.
Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010;11:1–15.
Malinsky M, Trucchi E, Lawson DJ, Falush D. RADpainter and fineRADstructure: population inference from RADseq data. Mol Biol Evol. 2018;35:1284–90.
Lawson DJ, Hellenthal G, Myers S, Falush D. Inference of population structure using dense haplotype data. PLOS Genet Public Library of Science. 2012;8:1–16.
Peakall R, Ruibal M, Lindenmayer DB, Url S. Spatial autocorrelation analysis offers new insights into gene flow in the Australian bush rat, Rattus fuscipes. Evolution (N Y). 2003;57:1182–95.
Vekemans X, Hardy OJ. New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol. 2004;13:921–35.
Peakall R, Smouse PE. GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–95.
Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180:977–93.
Narum SR, Hess JE. Comparison of FST outlier tests for SNP loci under selection. Mol Ecol Resour. 2011;11:184–94.
Pérez-Figueroa A, García-Pereira MJ, Saura M, Rolán-Alvarez E, Caballero A. Comparing three different methods to detect selective loci using dominant markers. J Evol Biol. 2010;23:2267–76.
Frichot E, Schoville SD, Bouchard G, François O. Testing for associations between loci and environmental gradients using latent factor mixed models. Mol Biol Evol. 2013;30:1687–99.
Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5.
Frichot E, François O. LEA: an R package for landscape and ecological association studies. Methods Ecol Evol. 2015;6:925–9.
Lamichhaney S, Fuentes-pardo AP, Rafati N, Ryman N, Mccracken GR. Parallel adaptive evolution of geographically distant herring populations on both sides of the North Atlantic Ocean. Proc Natl Acad Sci. 2017;114:3452–61.
van Zwieten PAM, Roest FC, Machiels MAM, van Densen WLT. Effects of inter-annual variability , seasonality and persistence on the perception of long-term trends in catch rates of the industrial pelagic purse-seine fishery of northern Lake Tanganyika (Burundi). Fish Res. 2002;54:329–48.
Phiri H, Shirakihara K. Distribution and seasonal movement of pelagic fish in southern Lake Tanganyika. Fish Res. 1999;41:63–71.
Sinclair MM, IIles TD. Population regulation and speciation in the oceans. J du Cons Int Explor la Mer. 1989;45:165–75.
Verheyen E, Rüber L, Snoeks J, Meyer A. Evolution on islands - Mitochondrial phylogeography of rock-dwelling cichlid fishes reveals evolutionary influence of historical lake level fluctuations of Lake Tanganyika, Africa. Philos Trans R Soc London Ser B Biol Sci. 1996;351:797 LP–805.
Koblmüller S, Salzburger W, Obermüller B, Eigner E, Sturmbauer C, Sefc KM. Separated by sand, fused by dropping water: habitat barriers and fluctuating water levels steer the evolution of rock-dwelling cichlid populations in Lake Tanganyika. Mol Ecol. 2011;20:2272–90.
Sturmbauer C, Baric S, Salzburger W, Rüber L, Verheyen E. Lake level fluctuations synchronize genetic divergences of cichlid fishes in African lakes. Mol Biol Evol. 2001;18:144–54.
Koblmüller S, Odhiambo EA, Sinyinza D, Sturmbauer C, Sefc KM. Big fish, little divergence: phylogeography of Lake Tanganyika’s giant cichlid, Boulengerochromis microlepis. Hydrobiologia. 2015;748:29–38.
Koblmüller S, Zangl L, Börger C, Daill D, Vanhove MPM, Sturmbauer C, et al. Only true pelagics mix: comparative phylogeography of Deepwater bathybatine cichlids from Lake Tanganyika. Hydrobiologia. 2018;3:1-11.
Whitehead PJP. FAO Species catalogue: Vol. 7 Clupeoid fishes of the world. FAO fish. synopsis. 1985.
Thompson AB. Simulation of reproductive rate, prey selection and the survival of pelagic fish of the African Great Lakes. Hydrobiologia. 1999;407:207–18.
Mulimbwa N, Shirakihara K. Growth, recruitment and reproduction of sardines (Stolothrissa tanganicae and Limnothrissa miodon) in northwester Lake Tanganyika. Tropics. 1994;4:57–67.
Kinsey ST, Orsoy T, Bert TM, Mahmoudi B. Population structure of the Spanish sardine Sardinella aurita: natural morphological variation in a genetically homogeneous population. Mar Biol. 1994;118:309–17.
García-Rodríguez FJ, García-Gasca SA, La C-AJD, Cota-Gómez VM. A study of the population structure of the Pacific sardine Sardinops sagax (Jenyns, 1842) in Mexico based on morphometric and genetic analyses. Fish Res. 2011;107:169–76.
Sebastian W, Sukumaran S, Zacharia PU, Gopalakrishnan A. Genetic population structure of Indian oil sardine, Sardinella longiceps assessed using microsatellite markers. Conserv Genet Springer Netherlands. 2017;18:951–64.
Atarhouch T, Rami M, Naciri M, Dakkak A. Genetic population structure of sardine (Sardina pilchardus) off Morocco detected with intron polymorphism (EPIC-PCR). Mar Biol. 2007;150:521–8.
Gonzalez EG, Zardoya R. Relative role of life-history traits and historical factors in shaping genetic population structure of sardines (Sardina pilchardus). BMC Evol Biol. 2007;7:197.
Limborg MT, Helyar SJ, De Bruyn M, Taylor MI, Nielsen EE, Ogden R, et al. Environmental selection on transcriptome-derived SNPs in a high gene flow marine fish, the Atlantic herring (Clupea harengus). Mol Ecol. 2012;21:3686–703.
Corander J, Majander KK, Cheng L, Merilä J. High degree of cryptic population differentiation in the Baltic Sea herring Clupea harengus. Mol Ecol. 2013;22:2931–40.
Mulimbwa N, Sarvala J, Micha J-C. The larval fishery on Limnothrissa miodon in the Congolese waters of Lake Tanganyika: impact on exploitable biomass and the value of the fishery. Fish Manag Ecol. 2018:1-7.
McLean KA, Byanaku A, Kubikonse A, Tshowe V, Katensi S, Lehman AG. Fishing with bed nets on Lake Tanganyika: a randomized survey. Malar J. 2014;13:395.
Acknowledgements
We thank Pierre-Denis Plisnier for his contribution to the research plan. We are grateful to all who helped us obtain clupeid samples: Fabrizia Ronco, Adrian Indermaur, Walter Salzburger, Donatien Muzumani Risasi, Fidel Muterezi Bukinga, Simon Kambale Mukeranya, Joseph Mbirize Ndalozibwa and Elias Bilali Mela. We thank the LBEG team for advice, discussion and support troughout the project. Michiel Jorissen contributed to the figures. We thank three anonymous reviewers for helping us improve the manuscript.
Funding
Research has been supported by the JEMU pilot project PopGenSprat (BELSPO funding) and the Belgian Development Cooperation through VLIR-UOS (VLADOC scholarship NDOC2016PR006 to ELRDK). MPMV was supported by the Belgian Directorate-General for Development Cooperation and Humanitarian Aid (CEBioS program). MPMV and NK are supported by the Czech Science Foundation (P505/12/G112 (ECIP)). These funding sources had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
Fish vouchers and tissue samples were deposited in the ichthyology collection of the Royal Museum for Central Africa (Tervuren, Belgium) under collection number 2016.20. Mitochondrial sequences were deposited in NCBI GenBank under accession numbers MH290064 - MH290159 and SNP data gained through RAD sequencing is submitted to Mendeley Data and can be retrieved via doi:https://doi.org/10.17632/hhd3mz3myd.1 .
Author information
Authors and Affiliations
Contributions
ELRDK wrote the manuscript and contributed to lab work and data analysis. ZDC performed lab work and analysis and contributed to the manuscript. Contribution of these two authors to the project is equal. NK, NM, PMM and MPMV carried out fieldwork. MVS, JAMR,FCFC, CV, MPMV and FAMV contributed to data analysis and data interpretation. NK and MV contributed to lab work. NK, CV, MV and NM contributed to interpretation of results and aided in writing the manuscript. MVS, JAMR, PMM and MPMV devised and oversaw the study, contributed to interpretation of results and critically revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Collection of specimens used in the study complied with institutional, national, and international guidelines. Fieldwork in D.R. Congo was carried out with the approval of the CRH – Uvira, which falls under the Congolese Ministry for science and technology (“Ministère National de la Recherche Scientifique et Technologie”) under mission statement 031/MINRST/CRH-U/2016. Samples were exported with an export permit from the CRH-Uvira. Sampling in Zambia was carried out with the approval of the Zambian Department of Fisheries under a memorandum of understanding between the University of Basel, the University of Zambia and the Zambian Department of Fisheries. An export permit was obtained from the Department of Fisheries at Mpulungu. Samples were imported in Belgium under import permit CONT/IEC/CHM/1365428 to the RMCA of the Belgian Federal Agency for the safety of the food chain (‘FAVV’). No animals were killed for this study, dead specimens were obtained from local fish markets.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Sequencing quality and information on missing data per individual. Table shows the sampling site, individual ID, number of SNPs, number of missing SNPs, frequency of missing SNPs, mean read depth and number of raw reads per individual. (PDF 370 kb)
Additional file 2:
Density plot of DAPC. Densities of individuals on the first discriminant function of the DAPC shown in Fig. 4. (PDF 28 kb)
Additional file 3:
Frequency distribution of global FST of Stolothrissa tanganicae per SNP. Grouping by sampling site and subbasin. (PDF 5 kb)
Additional file 4:
Outlier analysis based on BayeScan v2.1. A. FST-Log10 posterior probability for two levels (sampling site and subbasin) for each of the three replicates (R1, R2, R3). B. Q value for grouping according sampling site and subbasin with each of the three replicates (R1, R2, R3). (PDF 169 kb)
Additional file 5:
Individual-based latent fixed mixed model (LFMM) analysis. Distribution of adjusted p-values, corrected with the genomic inflation factor. Made with the LEA package in R. (PDF 4 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
De Keyzer, E.L.R., De Corte, Z., Van Steenberge, M. et al. First genomic study on Lake Tanganyika sprat Stolothrissa tanganicae: a lack of population structure calls for integrated management of this important fisheries target species. BMC Evol Biol 19, 6 (2019). https://doi.org/10.1186/s12862-018-1325-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12862-018-1325-8