Abstract
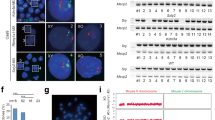
In modern molecular and cellular biology, CRISPR/Cas9 technology has been widely used for targeted modification of human and animal genomes. In this work, using molecular-cytogenetics methods, we have analyzed the karyotype in 18 mouse fibroblast cell lines (MFs), whose genome had a Cntn6 gene edited with CRISPR/Cas9. The modifications of Cntn6 gene were 2374-kb duplications, 1137-kb deletions, and inversions of similar size. In addition, cytogenetic analysis was performed for five control lines of mouse embryonic fibroblasts (MEFs) carrying an intact Cntn6 gene allele. The study showed the presence of a high level of polyploidy for MF lines that were heterozygous for Cntn6 gene inversions and homozygous and heterozygous for Cntn6 gene duplications (20–46%), as well as monosomy (1–9%) and trisomy (1–8%) for chromosome 6. It is important to note that no trisomy was detected in MEF lines carrying a deletion and duplication of Cntn6 gene in the compound, and the proportion of polyploid cells was minimal (1.5–5.7%). Thus, the data obtained indicate the destabilization of the karyotype of cell lines that underwent genome editing by CRISPR/Cas9.

Similar content being viewed by others
REFERENCES
Bolton, H., Graham, S.J.L., Niels, V.D.A., Kumar, P., Theunis, K., Gallardo, E.F., Voet, T., and Zernicka-Goetz, M., Mouse model of chromosome mosaicism reveals lineage-specific depletion of aneuploid cells and normal developmental potential, Nat. Commun., 2016, vol. 7, p. 11165.
Boroviak, K., Doe, B., Banerjee, R., Yang, F., and Bradley, A., Chromosome engineering in zygotes with CRISPR/Cas9, Genesis, 2016, vol. 54, p. 78.
Ca, L., Fisher, A.L., Huang, H., and Xie, Z., CRISPR-mediated genome editing and human diseases, Genes Dis., 2016, vol. 3, p. 244.
Canver, M.C., Bauer, D.E., Dass, A., Yien, Y.Y., Chung, J., Masuda, T., Maeda, T., Paw, B.H., and Orkin, S.H., Characterization of genomic deletion efficiency mediated by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells, J. Biol. Chem., 2017, vol. 292, p. 2556.
Cleveland, D.W., Mao, Y., and Sullivan, K.F., Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling, Cell, 2003, vol. 112, p. 407.
Cong, L., Ran, F.A., Cox, D., Lin, S., Barretto, R., Habib, N., Hsu, P.D., Wu, X., Jiang, W., Marraffini, L.A., and Zhang, F., Multiplex genome engineering using CRISPR/Cas systems, Science, 2013, vol. 339, p. 819.
Cox, D.B.T., Platt, R.D., and Zhang, F., Therapeutic genome editing: prospects and challenges, Nat. Ved., 2015, vol. 21, p. 121.
Crasta, K., Ganem, N.J., Dagher, R., Lantermann, A.B., Ivanova, E.V., Pan, Y., Nezi, L., Protopopov, A., Chowdhury, D., and Pellman, D., DNA breaks and chromosome pulverization from errors in mitosis, Nature, 2012, vol. 482, p. 53.
Fujii, W., Kawasaki, K., Sugiura, K., and Naito, K., Efficient generation of large-scale genome-modified mice using gRNA and Cas9 endonuclease, Nucleic Acids Res., 2013, vol. 41, p. e187.
Goepfert, T.M., McCarthy, M., Kittrell, F.S., Stephens, C., Ullrich, R.L., Brinkley, B.R., and Medina, D., Progesterone facilitates chromosome instability (aneuploidy) in p53 null normal mammary epithelial cells, FASEB J., 2000, vol. 14, p. 2221.
Hara, S., Kato, T., Goto, Y., Kubota, S., Tamano, M., Terao, M., and Takada, S., Microinjection-based generation of mutant mice with a double mutation and a 0.5 Mb deletion in their genome by the CRISPR/Cas9 system, J. Reprod. Dev., 2016, vol. 62, p. 531.
Hsu, P.D., Scott, D.A., Weinstein, J.A., Ran, F.A., Konermann, S., Agarwala, V., Li, Y., Fine, E.J., Wu, X., Shalem, O., Cradick, T.J., Marraffini, L.A., Bao, G., and Zhang, F., DNA targeting specificity of RNA-guided Cas9 nucleases, Nat. Biotechnol., 2013, vol. 31, p. 827.
Hsu, P.D., Lander, E.S., and Zhang, F., Development and applications of CRISPR-Cas9 for genome engineering, Cell, 2014, vol. 157, p. 1262.
Hussain, W., Mahmoodb, T., Hussain, J., Alid, N., Shahe, T., Qayyumf, S., and Khang, I., CRISPR/Cas system: a game changing genome editing technology, to treat human genetic diseases, Gene, 2019, vol. 685, p. 70.
Janssen, A., van der Burg, M., Szuhai, K., Kops, G.J., and Medema, R.H., Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations, Science, 2011, vol. 333, p. 1895.
Kalitsis, P., Fowler, K.J., Griffiths, B., Earle, E., Chow, C.W., Jamsen, K., and Choo, K.H.A., Increased chromosome instability but not cancer predisposition in haploinsufficient Bub3 mice, Genes Chromosomes Cancer, 2005, vol. 44, p. 29.
Korablev, A.N., Serova, I.A., and Serov, O.L., Generation of megabase-scale deletions, inversions and duplications involving the Contactin-6 gene in mice by CRISPR/Cas9 technology, BMC Genet., 2017, vol. 18, p. 112.
Kosicki, M., Tomberg, K., and Bradley, R., Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements, Nat. Biotechnol., 2018, vol. 36, p. 765.
Kraft, K., Geuer, S., Will, A.J., Chan, W.L., Paliou, C., Borschiwer, M., Harabula, I., Wittler, L., Franke, M., Ibrahim, D.M., Kragesteen, B.K., Spielmann, M., Mundlos, S., Lupianez, D.G., and Andrey, G., Deletions, inversions, duplications: engineering of structural variants using CRISPR/Cas in mice, Cell Rep., 2015, vol. 10, p. 833.
Lin, S.R., Yang, H.C., Kuo, Yi.T., Sung, K.C., Lin, Y.Y., Wang, H.Y., Wang, C.C., Shen, Y.C., Wu, F.Y., Kao, J.H., Chen, D.S., and Chen, P.J., The CRISPR/Cas9 system facilitates clearance of the intrahepatic HBV templates in vivo, Mol. Ther. Nucleic Acids, 2014, vol. 3, p. e186.
Liu, X., Wu, H., Loring, J., Hormuzdi, S., Disteche, C.M., Bornstein, P., and Jaenisch, R., Trisomy eight in ES cells is a common potential problem in gene targeting and interferes with germ line transmission, Dev. Dyn., 1997, vol. 209, p. 85.
Mehravar, M., Shirazi, A., Nazari, M., and Banan, M., Mosaicism in CRISPR/Cas9-mediated genome editing, Dev. Biol., 2019, vol. 445, p. 156.
Menzorov, A., Pristyazhnyuk, I., Kizilova, H., Yunusova, A., Battulin, N., Zhelezova, A., Golubitsa, A., and Serov, O.L., Cytogenetic analysis and Dlk1-Dio3 locus epigenetic status of mouse embryonic stem cells during early passages, Cytotechnology, 2016, vol. 68, p. 61.
Minina, Yu.M., Zhdanova, N.S., Shilov, A.G., Tolkunova, E.N., Liskovykh, M.A., andTomilin, A.N., Chromosomal instability of mouse pluripotent cells cultured in vitro, Cell Tissue Biol., 2010, vol. 4, p. 223.
Mollanoori, H., Shahraki, H., Rahmati, Y., and Teimourian, S., CRISPR/Cas9 and CAR-T cell, collaboration of two revolutionary technologies in cancer immunotherapy, an instruction for successful cancer treatment, Hum. Immunol., 2018, vol. 79, p. 876.
Pankowicz, F.P., Barzi, M., Legras, X., Hubert, L., Mi, T., Tomolonis, J.A., Ravishankar, M., Sun, Q., Yang, D., Borowiak, M., Sumazin, P., Elsea, S.H., Bissig-Choisat, B., and Bissig, K.D., Reprogramming metabolic pathways in vivo with CRISPR/Cas9 genome editing to treat hereditary tyrosinaemia, Nat. Commun., 2016, vol. 7, p. 1.
Podryadchikova, O.L., Pristyazhnyuk, I.E., Matveeva, N.M., and Serov, O.L., FISH analysis of regional replication of homologous chromosomes in hybrid cells obtained by fusion of embryonic stem cells with somatic cells, Tsitologiya, 2009, vol. 51, p. 500.
Pristyazhnyuk, I.E., Minina, J., Korablev, A., Serova, I., Fishman, V., Gridina, M., Rozhdestvensky, T.S., Gubar, L., Skryabin, B.V., and Serov, O.L., Time origin and structural analysis of the induced CRISPR/cas9 megabase-sized deletions and duplications involving the Cntn6 gene in mice, Sci. Rep., 2019, vol. 9, p. 14161.
Sakuma, T., Masaki, K., Abe-Chayama, H., Mochida, K., Yamamoto, T., and Chayama, K., Highly multiplexed CRISPR-Cas9-nuclease and Cas9-nickase vectors for inactivation of hepatitis B virus, Genes Cells, 2016, vol. 21, p. 1253.
Santaguida, S., Richardson, A., Iyer, D.R., M’Saad, O., Zasadil, L., Knouse, K.A., Wong, Y.L., Rhind, N., Desai, A., and Amon, A., Chromosome mis-segregation generates cell cycle-arrested cells with complex karyotypes that are eliminated by the immune system, Dev. Cell, 2017, vol. 41, p. 638.
Singla, S., Iwamoto-Stohl, L.K., Zhu, M., and Zernicka-Goetz, M., Autophagy-mediated apoptosis eliminates aneuploid cells in a mouse model of chromosome mosaicism, Nat. Commun., 2020, vol. 11, p. 2958.
Telenius, H., Pelmear, A.H., Tunnacliffe, A., Carter, N.P., Behmel, A., Ferguson-Smith, M.A., Nordenskjold, M., Pfragner, R., and Ponder, B.A., Cytogenetic analysis by chromosome painting using DOP-PCR amplified flow-sorted chromosomes, Genes Chromosomes Cancer, 1992, vol. 4, p. 226.
Thompson, S.L. and Compton, D.A., Chromosomes and cancer cells, Chromosome Res., 2011, vol. 19, p. 433.
Vetchinova, A.S., Simonova, V.V, Novosadova, E.V., Manuilova, E.S., Nenasheva, V.V., Tarantul, V.Z., Grivennikov, I.A., Khaspekov, L.G., and Illarioshkin, S.N., Cytogenetic analysis of the results of genome editing on the cell model of Parkinson’s disease, Bull. Exp. Biol. Med., 2018, vol. 165, p. 355.
Yang, E., O’Brien, P.C.M., and Ferguson-Smith, M.A., Comparative chromosome map of the laboratory mouse and Chinese hamster defined by reciprocal chromosome painting, Chromosome Res., 2000, vol. 8, p. 219.
Zhang, L., Jia, R., Palange, N.J., Satheka, A.C., Togo, J., An, Y., Humphrey, M., Ban, L., Ji, Y., Jin, H., Feng, X., and Zheng, Y., Large genomic fragment deletions and insertions in mouse using CRISPR/Cas9, PLoS One, 2015, vol. 10, p. e0120396.
Zhen, S., Lu, J.J., Wang, L.J., Sun, X.M., Zhang, J.Q., Li, X., Luo, W.J., and Zhao, L., In vitro and in vivo synergistic therapeutic effect of cisplatin with human papillomavirus16 E6/E7 CRISPR/Cas9 on cervical cancer cell line, Transl. Oncol., 2016, vol. 9, p. 498.
Funding
The authors declare that they have no conflicts of interest. This work was supported by the Russian Foundation for Basic Research (project no. 20-04-00463А) and budget project no.FWNR-2022-0019.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest. The authors declare that they have no conflicts of interest.
Statement on the welfare of animals. All procedures were carried out in accordance with an EU Council Regulation (November 24, 1966, 86/609/EEC) and approved by the Bioethics Commission of the Institute of Cytology and Genetics (authorization no. 24 of October 24, 2014).
Additional information
Translated by I. Fridlyanskaya
Abbreviations: BSA—bovine serum albumin; CRISPR/Cas9—clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9; FISH—fluorescent in situ hybridization; PCR—polymerase chain reaction; MEF—mouse embryonic fibroblast; MF—mouse fibroblast.
Rights and permissions
About this article
Cite this article
Minina, Y.M., Soroka, A.B., Karamysheva, T.V. et al. The Application of CRISPR/Cas9 Technology in Mice Zygotes to Obtain Duplications, Deletions and Inversions Affects Karyotype Stability. Cell Tiss. Biol. 17, 557–564 (2023). https://doi.org/10.1134/S1990519X23050085
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1990519X23050085