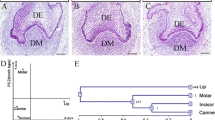
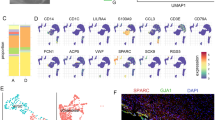
Abstract
In this study, we investigated the mechanism by which Msx1 and Pax9 affect tooth development in mice. The microarray data GSE32321, which contains Msx1 and Pax9 wildtype and knockout samples from mice oral epithelium (Epi) and dental mesenchymal (Mes) cells, was used to identify the differentially expressed genes (DEGs). The highest associated gene modules were then explored in the Epi-Msx, Epi-Pax, Mes-Msx, and Mes-Pax groups by weighted gene co-expression network analysis (WGCNA). Gene Ontology (GO) database analysis and hub gene screening were performed on the modules with the highest relevance. A total of 1467, 986, 1212, and 1293 DEGs were identified in the Epi-Msx, Epi-Pax, Mes-Msx, and Mes-Pax groups, respectively. Four highest associated gene modules were identified. GO enrichment analysis showed that the negative regulation of cell proliferation, cell adhesion, blood vessel development, and blood vessel morphogenesis was involved in tooth development. We further identified the hub genes IDH3A, SSPN, F13A1, and CBLN1, and found that gene expression values varied at different time points during tooth development. Moreover, IDH3A and CBLN1 were shown to be involved in oxidoreduction coenzyme metabolic processes and cell-cell adhesion. Overall, Msx1 and Pax9 play an important role in tooth development in mice, an effect which is probably associated with their regulation of IDH3A, SSPN, F13A1, and CBLN1.









Similar content being viewed by others
REFERENCES
Bei, M., Molecular genetics of tooth development, Curr. Opin. Genet. Dev., 2009, vol. 19, no. 5, pp. 504–510.
Benjamini, Y., Discovering the false discovery rate, J. R. Stat. Soc. B, 2010, vol. 72, no. 4, pp. 405–416.
Benjamini, Y., and Hochberg, Y., Controlling the false discovery rate: a practical and powerful approach to multiple testing, J. R. Stat. Soc. B, 1995, vol. 57, no. 1, pp. 289–300.
Chen, Y., Bei, M., Woo, I., et al., Msx1 controls inductive signaling in mammalian tooth morphogenesis, Development, 1996, vol. 122, no. 10, pp. 3035–3044.
Cho, S., Lee, J., Heo, J.S., et al., Gene expression change in human dental pulp cells exposed to a low-level toxic concentration of triethylene glycol dimethacrylate: an RNA-seq analysis, Basic Clin. Pharmacol., 2014, vol. 115, no. 3, pp. 282–290.
Demmer, R.T., Behle, J.H., Wolf, D.L., et al., Transcriptomes in healthy and diseased gingival tissues, J. Periodontol., 2008, vol. 79, no. 11, pp. 2112–2124.
Deza, M.M., and Deza, E., Encyclopedia of distances, in Encyclopedia of Distances, Berlin: Springer, 2009, pp. 1–583.
Estrada, E., Virtual identification of essential proteins within the protein interaction network of yeast, Proteomics, 2006, vol. 6, no. 1, pp. 35–40.
Falcon, S., and Gentleman, R., Using GOstats to test gene lists for GO term association, Bioinformatics, 2006, vol. 23, no. 2, pp. 257–258.
Findlay, A.S., Carter, R.N., Starbuck, B., et al., Mouse Idh3a mutations cause retinal degeneration and reduced mitochondrial function, Dis. Model Mech., 2018, vol. 11, no. 12, p. 036426.
Fujita, A., Sato, J.Ż.R., de Oliveira Rodrigues, L., et al., Evaluating different methods of microarray data normalization, BMC Bioinform., 2006, vol. 7, no. 1, p. 469.
Gentleman, R., Carey, V., Huber, W., et al., Bioinformatics and Computational Biology Solutions Using R and Bioconductor, NewYork: Springer Science and Business Media, 2006.
Jeong, H., Mason, S.P., Barabási, A.-L., et al., Lethality and centrality in protein networks, Nature, 2001, vol. 411, no. 6833, p. 41.
Jheon, A.H., Seidel, K., Biehs, B., et al., From molecules to mastication: the development and evolution of teeth, Wiley Interdiscip. Rev. Dev. Biol., 2013, vol. 2, no. 2, pp. 165–182.
Jia, Y., Nie, K., Li, J., et al., Identification of therapeutic targets for Alzheimer’s disease via differentially expressed gene and weighted gene co-expression network analyses, Mol. Med. Rep., 2016, vol. 14, no. 5, pp. 4844–4848.
Jussila, M., and Thesleff, I., Signaling networks regulating tooth organogenesis and regeneration, and the specification of dental mesenchymal and epithelial cell lineages, CSH Perspect. Biol., 2012, vol. 4, no. 4, p. a008425.
Kang, J., Bai, R., Liu, K., et al., Identification of significantly different modules between permanent and deciduous teeth by network and pathway analyses, Genet. Mol. Res., 2016, vol. 15, no. 4.
Kettunen, P.L., Sigbjørn, Furmanek, T., Fjeld, K., et al., Coordination of trigeminal axon navigation and patterning with tooth organ formation: epithelial-mesenchymal interactions, and epithelial Wnt4 and Tgfβ1 regulate semaphorin 3a expression in the dental mesenchyme, Development, 2005, vol. 132, no. 2, pp. 323–334.
Langfelder, P. and Horvath, S., WGCNA: an R package for weighted correlation network analysis, BMC Bioinformatics, 2008, vol. 9, no. 1, p. 559.
Lesot, H. and Brook, A., Epithelial histogenesis during tooth development, Arch. Oral Biol., 2009, vol. 54, no.1, pp. S25–S33.
Li, Z., Yu, M., and Tian, W., An inductive signalling network regulates mammalian tooth morphogenesis with implications for tooth regeneration, Cell Proliferation, 2013, vol. 46, no. 5, pp. 501–508.
Lumsden, A., Spatial organization of the epithelium and the role of neural crest cells in the initiation of the mammalian tooth germ, Development, 1988, vol. 103, suppl., pp. 155–169.
O’Connell, D.J., Ho, J.W., Mammoto, T., et al., A Wnt-bmp feedback circuit controls intertissue signaling dynamics in tooth organogenesis, Sci. Signal., 2012, vol. 5, no. 206, pp. ra4–ra4.
Oldham, M.C., Konopka, G., Iwamoto, K., et al., Functional organization of the transcriptome in human brain, Nat. Neurosci., 2008, vol. 11, no. 11, p. 1271.
Peters, H., Neubüser, A., Kratochwil, K., et al., Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities, Gene Dev., 1998, vol. 12, no. 17, pp. 2735–2747.
Plaisier, C.L., Horvath, S., Huertas-Vazquez, A., et al., A systems genetics approach implicates USF1, FADS3, and other causal candidate genes for familial combined hyperlipidemia, PLoS Genet., 2009, vol. 5, no. 9. e1000642.
Press, W., Teukolsky, S., Vetterling, W., et al., Section 16.4. Hierarchical clustering by phylogenetic trees, in Numerical Recipes: The Art of Scientific Computing, NewYork: Cambridge Univ. Press, 2007.
Rostampour, N., Linking New Genes to an Odontogenesis TP63-Mediated Gene Regulatory Network That Is Peripheral to Jaw Morphogenesis, Saskatoon: University of Saskatchewan, 2018.
Satokata, I. and Maas, R., Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development, Nat. Genet., 1994, vol. 6, no. 4, p. 348.
Saxén, L. and Thesleff, I., Epithelial–mesenchymal interactions in murine organogenesis, Ciba Found. Symp., 1992, vol. 165, pp. 183–193.
Smoot, M.E., Ono, K., Ruscheinski, J., et al., Cytoscape 2.8: new features for data integration and network visualization, Bioinformatics, 2010, vol. 27, no. 3, pp. 431–432.
Szekely, G.J., and Rizzo, M.L., Hierarchical clustering via joint between-within distances: Extending Ward’s minimum variance method, J. Classif., 2005, vol. 22, no. 2, pp. 151–183.
Thesleff, I., The genetic basis of normal and abnormal craniofacial development, Acta Odontol. Scand., 1998, vol. 56, no. 6, pp. 321–325.
Thesleff, I., Epithelial–mesenchymal signalling regulating tooth morphogenesis, J. Cell Sci, 2003, vol. 116, no. 9, pp. 1647–1648.
Troyanskaya, O., Cantor, M., Sherlock, G., et al., Missing value estimation methods for DNA microarrays, Bioinformatics, 2001, vol. 17, no. 6, pp. 520–525.
Tucker, A.S., Matthews, K.L., and Sharpe, P.T., Transformation of tooth type induced by inhibition of BMP signaling, Science, 1998, vol. 282, no. 5391, pp. 1136–1138.
Wang, L., Cao, C., Ma, Q., et al., RNA-seq analyses of multiple meristems of soybean: novel and alternative transcripts, evolutionary and functional implications, BMC Plant Biol., 2014, vol. 14, no. 1, p. 169.
Yoneda, T., Tomofuji, T., Kawabata, Y., et al., Application of coenzyme Q10 for accelerating soft tissue wound healing after tooth extraction in rats, Nutrients, 2014, vol. 6, no. 12, pp. 5756–5769.
Funding
None.
Author information
Authors and Affiliations
Contributions
Conception and design of the research: FW and WJ; acquisition of data: FW and WJ; analysis and interpretation of data: RL and BC; statistical analysis: RL and BC; drafting the manuscript: FW and WJ; revision of manuscript for important intellectual content: RL and BC. All authors have read and approved the final manuscript FW and WJ contributed equally to this work.
Corresponding authors
Ethics declarations
COMPLIANCE WITH ETHICAL STANDARDS
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
AVAILABILITY OF DATA AND MATERIAL
The data used to support the findings of this study are available from the corresponding author upon request.
ADDITION INFORMATION
Feng Wang and Wen Jiang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Feng Wang, Jiang, W., Chen, B. et al. The Mechanism of MSX1 and PAX9 Implication in Tooth Development Based on the Weighted Gene Co-Expression Network Analysis. Russ J Dev Biol 52, 187–198 (2021). https://doi.org/10.1134/S1062360421030085
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1062360421030085