Abstract
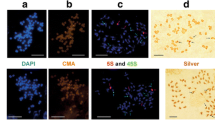
Using sequence-tagged DNA sequencing on the Roche 454 platform, we studied intragenomic polymorphism of one of the 35S rRNA regions (18S rDNA fragment–ITS1–fragment of 5.8S rDNA) in three hexaploid Avena species with karyotypes AACCDD and a tetraploid species A. insularis (AAСС or CCDD). Instead of expected 50% of C-variant ITS1 in A. insularis and 33% of C-variant ITS1 in hexaploids A. fatua, A. ludoviciana, and A. sterilis, the actual rate of C-subgenome specific ITSs comprised around 3.3% of rDNA in A. insularis and 1.4–2.4% of rDNA in hexaploid genomes. The 18S rDNA (fragment), ITS1 and 5.8S rDNA (small fragment) of the C-subgenome origin were 10 times more variable than the same sequences from the A-genome. Some of the C-subgenome sequences contained deletions, including deletions in the 18S rRNA coding region. The results of FISH hybridization with pTa71 and pTa794 confirm the fact that polyploids lost a significant part of the 35S rDNA and 5S rDNA obtained from a diploid ancestor with the CC karyotype. Our results show that the loss of the 35S rDNA of the C type occurs against the background of multiple single nucleotide polymorphisms (SNPs) and deletions accumulation in these sequences. The fact that all C-subgenome ITS1 sequences in the genomes of polyploids were represented by single (unique) copies might indicate that the appearance of multiple mutations in the “repressed” 35S rRNA loci was not accompanied by homogenization of rDNA. Hence, there is a reason to believe that the process of rDNA isogenization and the process of transcription/silencing are related phenomena.



Similar content being viewed by others
REFERENCES
Van de Peer, Y., Mizrachi, E., and Marchal, K., The evolutionary significance of polyploidy, Nat. Rev. Genet., 2017, vol. 18, pp. 411—424. https://doi.org/10.1038/nrg.2017.26
Soltis, D.E., Visger, C.J., Marchant, D.B., and Soltis, P.S., Polyploidy: pitfalls and paths to a paradigm, Am. J. Bot., 2016, vol. 103, pp. 1146—1166. https://doi.org/10.3732/ajb.1500501
Hu, G. and Wendel, J.F., Cis–trans controls and regulatory novelty accompanying allopolyploidization, New Phytol., 2019, vol. 221, pp. 1691—1700. https://doi.org/10.1111/nph.15515
Rodionov, A.V., Amosova, A.V., Belyakov, E.A., et al., Genetic consequences of interspecific hybridization, its role in speciation and phenotypic diversity of plants, Russ. J. Genet., 2019, vol. 55, no. 3, pp. 278—294. https://doi.org/10.1134/S1022795419030141
Rodionov, A.V., Interspecific hybridization and polyploidy in the evolution of plants, Vavilovskii Zh. Genet. Sel., 2013, vol. 17, no. 4 (2), pp. 916—929.
Rodionov, A.V., Nosov, N.N., Kim, E.S., et al., The origin of polyploid genomes of bluegrasses Poa L. and gene flow between northern pacific and sub-Antarctic islands, Russ. J. Genet., 2010, vol. 46, no. 12, pp. 1407—1416.
Xiong, Z., Gaeta, R.T., and Pires, J.C., Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus,Proc. Natl. Acad. Sci. U.S.A., 2011, vol. 108, pp. 7908—7913. https://doi.org/10.1073/pnas.1014138108
Lipman, M.J., Chester, M., Soltis, P.S., and Soltis, D.E., Natural hybrids between Tragopogon mirus and T. miscellus (Asteraceae): a new perspective on karyotypic changes following hybridization at the polyploid level, Am. J. Bot., 2013, vol. 100, pp. 2016—2022. https://doi.org/10.3732/ajb.1300036
Navashin, M., Chromosomal alterations caused by hybridization and their bearing upon certain general genetic problems, Cytologia, 1934, vol. 5, pp. 169—203.
Kovarik, A., Dadejova, M., Lim, Y.K., et al., Evolution of rDNA in Nicotiana allopolyploids: a potential link between rDNA homogenization and epigenetics, Ann. Bot., 2008, vol. 101, pp. 815—823. https://doi.org/10.1093/aob/mcn019
Matyášek, R., Renny-Byfield, S., Fulneček, J., et al., Next generation sequencing analysis reveals a relationship between rDNA unit diversity and locus number in Nicotiana diploids, BMC Genomics, 2012, vol. 13, p. 722. https://doi.org/10.1186/1471-2164-13-722
Long, H., Chen, C., Wang, B., and Feng, Y., rDNA genetic imbalance and nucleolar chromatin restructuring is induced by distant hybridization between Raphanus sativus and Brassica alboglabra,PLoS One, 2015, vol. 10, no. 2. e0117198. https://doi.org/10.1371/journal.pone.0117198
Punina, E.O., Machs, E.M., Krapivskaya, E.E., et al., Interspecific hybridization in the genus Paeonia (Paeoniaceae): polymorphic sites in transcribed spacers of the 45S rRNA genes as indicators of natural and artificial peony hybrids, Russ. J. Genet., 2012, vol. 48, no. 7, pp. 684—697. https://doi.org/10.1134/S1022795412070113
Rodionov, A.V., Dobryakova, K.S., and Punina, E.O., Polymorphic sites in ITS1–5.8S rDNA–ITS2 region in hybridogenic genus × Elyhordeum and putative interspecific hybrids Elymus (Poaceae: Triticeae), Russ. J. Genet., 2018, vol. 54, no. 9, pp. 999—1014. https://doi.org/10.1134/S1022795418090120
Rodionov, A.V., Gnutikov, A.A., Kotsinyan, A.R., et al., ITS1–5.8S rDNA–ITS2 sequence in 35S rRNA genes as marker for reconstruction of phylogeny of grasses (Poaceae family), Biol. Bull. Rev., 2017, vol. 7, no. 2, pp. 85—102. https://doi.org/10.1134/S2079086417020062
Rodionov, A.V., Tyupa, N.B., Kim, E.S., et al., Genomic configuration of the autotetraploid oat species Avena macrostachya inferred from comparative analysis of ITS1 and ITS2 sequences: on the oat karyotype evolution during the early events of the Avena species divergence, Russ. J. Genet., 2005, vol. 41, no. 5, pp. 518—528. https://doi.org/10.1007/s11177-005-0120-y
Nikoloudakis, N. and Katsiotis, A., The origin of the C-genome and cytoplasm of Avena polyploids, Theor. Appl. Genet., 2008, vol. 117, pp. 273—281. https://doi.org/10.1007/s00122-008-0772-9
Peng, Y.Y., Baum, B.R., Ren, C.Z., et al., The evolution pattern of rDNA ITS in Avena and phylogenetic relationship of the Avena species (Poaceae: Aveneae), Hereditas, 2010, vol. 147, pp. 183—204. https://doi.org/10.1111/j.1601-5223.2010.02172.x
Jellen, E.N., Gill, B.S., and Cox, T.S., Genomic in situ hybridization differentiates between A/D-and C-genome chromatin and detects intergenomic translocations in polyploid oat species (genus Avena), Genome, 1994, vol. 37, pp. 613—618.
Irigoyen, M.L., Loarce, Y., Linares, C., et al., Discrimination of the closely related A and B genomes in AABB tetraploid species of Avena,Theor. Appl. Genet., 2001, vol. 103, pp. 1160—1166.
Shelukhina, O., Badaeva, E., Loskutov, I., and Pukhal’sky, V.A., A comparative cytogenetic study of the tetraploid oat species with the A and C genomes: Avena insularis, A. magna, and A. murphyi,Russ. J. Genet., 2007, vol. 43, pp. 613—626.
Badaeva, E.D., Shelukhina, O.Y., Diederichsen, A., et al., Comparative cytogenetic analysis of Avena macrostachya and diploid C-genome Avena species, Genome, 2010, vol. 53, pp. 125—137. https://doi.org/10.1139/g09-089
Tyupa, N.B., Kim, E.S., Loskutov, I.G., and Rodionov, A.V., To the origin of polyploids in the genus Avena L.: molecular phylogenetic research, Tr. Prikl. Bot.,Genet. Sel., 2009, vol. 165, pp. 13—20.
Fominaya, A., Loarce, Y., Montes, A., and Ferrer, E., Chromosomal distribution patterns of the (AC) 10 microsatellite and other repetitive sequences, and their use in chromosome rearrangement analysis of species of the genus Avena,Genome, 2016, vol. 60, pp. 216—227. https://doi.org/10.1139/gen-2016-0146
Rajhathy, T. and Thomas, H.T., Chromosomal differentiation and speciation in diploid Avena,Can. J. Genet. Cytol., 1967, vol. 9, pp. 52—68.
Doyle, J.J. and Doyle, J.L., A rapid DNA isolation procedure for small quantities of fresh leaf tissue, Phytochem. Bull., 1987, vol. 19, pp. 11—15.
Ridgway, K.P., Duck, J.M., and Young, J.P.W., Identification of roots from grass swards using PCR-RFLP and FFLP of the plastid trnL (UAA) intron, BMC Ecol., 2003, vol. 3, no. 8. https://doi.org/10.1186/1472-6785-3-8
Stanford, A.M., Harden, R., and Parks, C.R., Phylogeny and biogeography of Juglans (Juglandaceae) based on matK and ITS sequence data, Am. J. Bot., 2000, vol. 87, pp. 872—882.
White, T.J., Bruns, T., Lee, S., and Taylor, J., Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, PCR Protoc.:Guide Methods Appl., 1990, vol. 18, pp. 315—322.
Bolger, A.M., Lohse, M., and Usadel, B., Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics, 2014, vol. 30, pp. 2114—2120. https://doi.org/10.1093/bioinformatics/btu170
Aronesty, E., Comparison of sequencing utility programs, Open Bioinf. J., 2013, vol. 7, no. 1. https://doi.org/10.2174/18750362013070100011
Okonechnikov, K., Golosova, O., Fursov, M., et al., Unipro UGENE: a unified bioinformatics toolkit, Bioinformatics, 2012, vol. 28, pp. 1166—1167.https://doi.org/10.1093/bioinformatics/bts091
Kumar, S., Stecher, G., and Tamura, K., MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets, Mol. Biol. Evol., 2016, vol. 33, pp. 1870—1874. https://doi.org/10.1093/molbev/msw054
Clement, M., Posada, D.C.K.A., and Crandall, K.A., TCS: a computer program to estimate gene genealogies, Mol. Ecol., 2000, vol. 9, pp. 1657—1659.
Múrias dos Santos, A., Cabezas, M.P., Tavares, A.I., et al., tcsBU: a tool to extend TCS network layout and visualization, Bioinformatics, 2015, vol. 32, pp. 627—628. https://doi.org/10.1093/bioinformatics/btv636
Badaeva, E.D., Badaev, N.S., Gill, B.S., and Filatenko, A.A., Intraspecific karyotype divergence in Triticum araraticum (Poaceae), Plant Syst. Evol., 1994, vol. 192, pp. 117—145.https://doi.org/10.1007/BF00985912
Amosova, A.V., Bolsheva, N.L., Samatadze, T.E., et al., Molecular cytogenetic analysis of Deschampsia antarctica Desv. (Poaceae), Maritime Antarctic, PLoS One, 2015, vol. 10, no. 9. e0138878. https://doi.org/10.1371/journal.pone.0138878
Gerlach, W.L. and Bedbrook, J.R., Cloning and characterization of ribosomal RNA genes from wheat and barley, Nucleic Acids Res., 1979, vol. 7, pp. 1869—1885. PMID: 537913.https://doi.org/10.1093/nar/7.7.1869
Gerlach, W.L. and Dyer, T.A., Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes, Nucleic Acids Res., 1980, vol. 8, pp. 4851—4855.
Muravenko, O.V., Yurkevich, O.Y., Bolsheva, N.L., et al., Comparison of genomes of eight species of sections Linum and Adenolinum from the genus Linum based on chromosome banding, molecular markers and RAPD analysis, Genetica, 2009, vol. 135, pp. 245—255. https://doi.org/10.1007/s10709-008-9273-7
Loskutov, I.G., Interspecific crosses in the genus Avena L., Russ. J. Genet., 2001, vol. 37, no. 5, pp. 467—475. https://doi.org/10.1023/A:1016697812009
Ladizinsky, G., A new species of Avena from Sicily, possibly the tetraploid progenitor of hexaploid oats, Genet. Res. Crop Evol., 1998, vol. 45, pp. 263—269.
Jellen, E.N. and Ladizinsky, G., Giemsa C-banding in Avena insularis Ladizinsky, Genet. Res. Crop Evol., 2000, vol. 47, pp. 227—230.
Zhou, X., Jellen, E.N., and Murphy, J.P., Progenitor germplasm of domesticated hexaploid oat, Crop Sci., 1999, vol. 39, pp. 1208—1214.
Loskutov, I.G., On evolutionary pathways of Avena species, Genet. Res. Crop Evol., 2008, vol. 55, pp. 211—220.
Irigoyen, M.L., Linares, C., Ferrer, E., and Fominaya, A., Fluorescence in situ hybridization mapping of Avena sativa L. cv. SunII and its monosomic lines using cloned repetitive DNA sequences, Genome, 2002, vol. 45, pp. 1230—1237.
Murai, K. and Tsunewaki, K., Chloroplast genome evolution in the genus Avena,Genetics, 1987, vol. 116, pp. 613—621.
Rines, H.W., Gengenbach, B.G., Boylan, K.L., and Storey, K.K., Mitochondrial DNA diversity in oat cultivars and species, Crop Sci., 1988, vol. 28, pp. 171—176.
Mogensen, H.L., The hows and whys of cytoplasmic inheritance in seed plants, Am. J. Bot., 1996, vol. 83, pp. 383–404.
Peng, Y.Y., Wei, Y.M., Baum, B.R., et al., Phylogenetic investigation of Avena diploid species and the maternal genome donor of Avena polyploids, Taxon, 2010, vol. 59, pp. 1472—1482.
Fu, Y.B., Oat evolution revealed in the maternal lineages of 25 Avena species, Sci. Rep., 2018, vol. 8, no. 1, p. 4252. https://doi.org/10.1038/s41598-018-22478-4
Kovarik, A., Matyasek, R., Lim, K.Y., et al., Concerted evolution of 18–5.8–26S rDNA repeats in Nicotiana allotetraploids, Biol. J. Linn. Soc., 2004, vol. 82, pp. 615—625.
Ganley, A.R. and Kobayashi, T., Highly efficient concerted evolution in the ribosomal DNA repeats: total rDNA repeat variation revealed by whole-genome shotgun sequence data, Genome Res., 2007, vol. 17, pp. 184—191.
Eickbush, T.H. and Eickbush, D.G., Finely orchestrated movements: evolution of the ribosomal RNA genes, Genetics, 2007, vol. 175, pp. 477—485.
Dadejová, M., Lim, K.Y., Soucková-Skalická, K., et al., Transcription activity of rRNA genes correlates with a tendency towards intergenomic homogenization in Nicotiana allotetraploids, New Phytol., 2007, vol. 174, pp. 658—668.
Sochorová, J., Coriton, O., Kuderová, A., et al., Gene conversion events and variable degree of homogenization of rDNA loci in cultivars of Brassica napus,Ann. Bot., 2016, vol. 119, pp. 13—26. https://doi.org/10.1093/aob/mcw187
Lunerová, J., Renny-Byfield, S., Matyášek, R., et al., Concerted evolution rapidly eliminates sequence variation in rDNA coding regions but not in intergenic spacers in Nicotiana tabacum allotetraploid, Plant Syst. Evol., 2017, vol. 303, pp. 1043—1060.
Peng, J.C. and Karpen, G.H., H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability, Nat. Cell Biol., 2007, vol. 9, pp. 25—35.
Gaillard, H. and Aguilera, A., Transcription as a threat to genome integrity, Ann. Rev. Biochem., 2016, vol. 85, pp. 291—317. https://doi.org/10.1146/annurev-biochem-060815-014908
Okamoto, H., Watanabe, T.A., and Horiuchi, T., Double rolling circle replication (DRCR) is recombinogenic, Genes Cells, 2011, vol. 16, pp. 503—513.
Noto, T., Kataoka, K., Suhren, J.H., et al., Small-RNA-mediated genome-wide trans-recognition network in Tetrahymena DNA elimination, Mol. Cell, 2015, vol. 59, pp. 229—242. https://doi.org/10.1016/j.molcel.2015.05.024
Funding
This work was supported by the Russian Foundation for Basic Research, project no. 17-00-00340 (17-00-00336, 17-00-00337, 17-00-00338).
Author information
Authors and Affiliations
Contributions
Both authors contributed equally to this work and are listed in alphabetical order.
Corresponding author
Ethics declarations
The authors declare no conflict of interest. This article does not contain any studies carried out with humans or animals as objects.
Additional information
Translated by M. Bibov
Rights and permissions
About this article
Cite this article
Rodionov, A.V., Amosova, A.V., Krainova, L.M. et al. Phenomenon of Multiple Mutations in the 35S rRNA Genes of the C Subgenome of Polyploid Avena L.. Russ J Genet 56, 674–683 (2020). https://doi.org/10.1134/S1022795420060095
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795420060095