Abstract
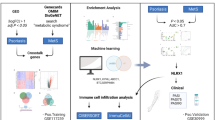
IQGAP proteins coordinate cell signaling cascades acting as scaffolds for assembly of multiprotein complexes. They mediate growth factor signaling, epithelial-mesenchymal transition, cell proliferation and migration. All of these pathways are important for the pathogenesis of psoriasis, so we suggested that the genes of the IQGAP family may play important roles in the development of this disease. qPCR analysis of the IQGAP1, IQGAP2, and IQGAP3 expression in skin of psoriasis patients showed the genes to be differentially expressed, with IQGAP1 being downregulated and IQGAP3 upregulated. Next we conducted a bioinformatic analysis of IQGAP protein partners in order to identify the protein interactions which would explain the difference between the lesional and visually nonlesional psoriatic skin. On the basis of the meta-analysis of the RNAseq data and the protein-protein interaction databases, we constructed IQGAP PPI graphs which highlighted the IQGAP protein partners involved in psoriasis. Therefore, our studies confirmed the hypothesis on the role of the IQGAP family in the pathogenesis of psoriasis.


Similar content being viewed by others
REFERENCES
Di Meglio, P., Perera, G.K., and Nestle, F.O., The multitasking organ: recent insights into skin immune function, Immunity, 2011, vol. 35, no. 6, pp. 857—869. https://doi.org/10.1016/j.immuni.2011.12.003
Ganguly, K., Upadhyay, S., Irmler, M., and Takenaka, S., Pathway focused protein profiling indicates differential function for IL-1B, -18 and VEGF during initiation and resolution of lung inflammation evoked by carbon nanoparticle exposure in mice, Part. Fibre Toxicol., 2009, vol. 6, no. 1, p. 31. https://doi.org/10.1186/1743-8977-6-31
Lande, R., Gregorio, J., Facchinetti, V., et al., Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide, Nature, 2007, vol. 449, no. 7162, p. 564. https://doi.org/10.1038/nature06116
Capon, F., The genetic basis of psoriasis, J. Mol. Sci., 2017, vol. 18, no. 12, pii: E2526. https://doi.org/10.3390/ijms18122526
Monteleon, C.L., McNeal, A., Duperret, E.K., et al., IQGAP1 and IQGAP3 serve individually essential roles in normal epidermal homeostasis and tumor progression, J. Invest. Dermatol., 2015, vol. 135, no. 9, pp. 2258—2265. https://doi.org/10.1016/j.cellsig.2009.02.023
Johnson, M., Sharma, M., and Henderson, B.R., IQGAP1 regulation and roles in cancer, Cell. Signalling, 2009, vol. 21, no. 10, pp. 1471—1478.
Vaitheesvaran, B., Hartil, K., Navare, A., et al., Role of the tumor suppressor IQGAP2 in metabolic homeostasis: зossible link between diabetes and cancer, Metabolomics, 2014, vol. 10, no. 5, pp. 920—937. https://doi.org/10.1007/s11306-014-0639-9
Kumar, D., Hassan, M.K., Pattnaik, N., Reduced expression of IQGAP2 and higher expression of IQGAP3 correlates with poor prognosis in cancers, PLoS One, 2017, vol. 12, no. 10. e0186977. https://doi.org/10.1371/journal.pone.0186977
Xie, Y., Zheng, L., and Tao, L., Downregulation of IQGAP2 correlates with prostate cancer recurrence and metastasis, Transl. Oncol., 2019, vol. 12, no. 2, pp. 236—244. https://doi.org/10.1016/j.tranon.2018.10.009
Hu, G., Xu, Y., Chen, W., et al., RNA interference of IQ motif containing GTPase-activating protein 3 (IQGAP3) inhibits cell proliferation and invasion in breast carcinoma cells, Oncol. Res., 2016, vol. 24, no. 6, pp. 455—461. https://doi.org/10.3727/096504016X14685034103635
Wang, S., Watanabe, T., Noritake, J., et al., IQGAP3, a novel effector of Rac1 and Cdc42, regulates neurite outgrowth, J. Cell Sci., 2007, vol. 120, no. 4, pp. 567—577. https://doi.org/10.1242/jcs.03356
McNulty, D.E., Li, Z., White, C.D., et al., Map kinase scaffold IQGAP1 binds the EGF receptor and modulates its activation, J. Biol. Chem., 2011, pp. 15010—15021. https://doi.org/10.1074/jbc.M111.227694
White, C.D., Erdemir, H.H., and Sacks, D.B., IQGAP1 and its binding proteins control diverse biological functions, Cell. Signalling, 2012, vol. 24, no. 4, pp. 826—834. https://doi.org/10.1016/j.cellsig.2011.12.005
Watanabe, T., Wang, S., and Kaibuchi, K., IQGAPs as key regulators of actin—cytoskeleton dynamics, Cell Struct. Funct., 2015, vol. 40, no. 2, pp. 69—77. https://doi.org/10.1247/csf.15003
Livak, K.J. and Schmittgen, T.D., Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method, Methods, 2001, vol. 25, no. 4, pp. 402—408. https://doi.org/10.1006/meth.2001.1262
Cock, P.J.A., Fields, C.J., Goto, N., The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants, Nucleic Acids Res., 2009, vol. 38, no. 6, pp. 1767—1771. https://doi.org/10.1093/nar/gkp1137
Bolger, A.M., Lohse, M., and Usadel, B., Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics, 2014, vol. 30, no. 15, pp. 2114—2120. https://doi.org/10.1093/bioinformatics/btu170
Dobin, A., Davis, C.A., Schlesinger, F., and Drenkow, J., STAR: ultrafast universal RNA-seq aligner, Bioinformatics, 2013, vol. 29, no. 1, pp. 15—21. https://doi.org/10.1093/bioinformatics/bts635
Szklarczyk, D., Franceschini, A., Kuhn, M., et al., The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored, Nucleic Acids Res., 2010, vol. 39, suppl. 1, pp. D561—D568. https://doi.org/10.1093/nar/gkq973
Han, W.S., Lee, J., Pham, M.D., and Yu, J.X., iGraph: a framework for comparisons of disk-based graph indexing techniques, Proc. VLDB Endowment, 2010, vol. 3. nos. 1—2, pp. 449—459. https://doi.org/10.14778/1920841.1920901
Chuang, H.Y., Lee, E., Liu, Y.T., et al., Network-based classification of breast cancer metastasis, Mol. Syst. Biol., 2007, vol. 3, no. 1, p. 140. https://doi.org/10.1038/msb4100180
Skovorodnikova, P.A., Chesnokov, M.S., Budko, A.A., et al., Scaffold proteins of the IQGAP family—multifunctional regulators of intracellular signaling and tumor transformation, Usp. Mol. Onkol., 2017, vol. 4, no. 2.
Hedman, A.C., Smith, J.M., and Sacks, D.B., The biology of IQGAP proteins: beyond the cytoskeleton, EMBO Rep., 2015, vol. 16, no. 4, pp. 427—446.
Buttrick, T.S., Wang, W., Yung, C., et al., Foxo1 promotes Th9 cell differentiation and airway allergy, Sci. Rep., 2018, vol. 8, no. 1, p. 818. https://doi.org/10.4049/jimmunol.1500849
Eberle, F.C., Bruck, J., Holstein, J., et al., Recent advances in understanding psoriasis version 1, F1000Res, 2016, vol. 5. https://doi.org/10.12688/f1000research.7927.1
Pandey, D., Berkowitz, D., and Romer, L., Kruppel-like factor 15: a critical transcriptional regulator of hypoxia induced endothelial arginase 2, FASEB J., 2016, vol. 30, no. 11, suppl. 1.
Mehta, N.N., Shin, D.B., Joshi, A.A., et al., Effect of 2 psoriasis treatments on vascular inflammation and novel inflammatory cardiovascular biomarkers: a randomized placebo-controlled trial, Circ. Cardiovasc. Imaging, 2018, vol. 11, no. 6. e007394. https://doi.org/10.1161/CIRCIMAGING.117.007394
Jain, M.K., Sangwung, P., and Hamik, A., Regulation of an inflammatory disease: Krüppel-like factors and atherosclerosis, Arterioscler. Thromb. Vasc. Biol., 2014, vol. 34, no. 3, pp. 499—508. https://doi.org/10.1161/ATVBAHA.113.301925
Dubrac, S. and Schmuth, M., PPAR-alpha in cutaneous inflammation, Dermatoendocrinology, 2011, vol. 3, no. 1, pp. 23—26. https://doi.org/10.4161/derm.3.1.14615
Carrera, M., Bitu, C.C., de Oliveira, C.E., et al., HOXA10 controls proliferation, migration and invasion in oral squamous cell carcinoma, Int. J. Clin. Exp. Pathol., 2015, vol. 8, no. 4, p. 3613.
Zolotarenko, A., Chekalin, E., Piruzian, E., and Bruskin, S., FRA1 mediates the activation of keratinocytes: implications for the development of psoriatic plaques, Biochim. Biophys. Acta Mol. Basis Dis., 2018, vol. 1864, no. 12, pp. 3726—3734. https://doi.org/10.1016/j.bbadis.2018.09.016
Brandner, J.M., Zorn-Kruppa, M., Yoshida, T., et al., Epidermal tight junctions in health and disease, Tissue Barriers, 2015, vol. 3, nos. 1—2. e974451. https://doi.org/10.4161/21688370.2014.974451
Collin, M., Dickinson, R., and Bigley, V., Haematopoietic and immune defects associated with GATA2 mutation, Br. J. Haematol., 2015, vol. 169, no. 2, pp. 173—187. https://doi.org/10.1111/bjh.13317
Qasim, M., Rahman, H., Oellerich, M., and Asif, A.R., Differential proteome analysis of human embryonic kidney cell line (HEK-293) following mycophenolic acid treatment, Proteome Sci., 2011, vol. 9, no. 1, p. 57. https://doi.org/10.1161/01.ATV.0000126373.52450.32
Kunimoto, K., Nojima, H., Yamazaki, Y., and Yoshikawa, T., Involvement of IQGAP3, a regulator of Ras/ERK-related cascade, in hepatocyte proliferation in mouse liver regeneration and development, J. Cell. Physiol., 2009, vol. 220, no. 3, pp. 621—631. https://doi.org/10.1002/jcp.21798
Yang, Y., Zhao, W., Xu, Q.W., et al., IQGAP3 promotes EGFR-ERK signaling and the growth and metastasis of lung cancer cells, PLoS One, 2014, vol. 9, no. 5. e97578. https://doi.org/10.1371/journal.pone.0097578
Shi, Y., Qin, N., Zhou, Q., et al., Role of IQGAP3 in metastasis and epithelial–mesenchymal transition in human hepatocellular carcinoma, J. Translat. Med., 2017, vol. 15, no. 1, p. 176. https://doi.org/10.1186/s12967-017-1275-8
Funding
This study was funded by a grant from the Russian Science Foundation (project no. 18-75-00126, supervisor A.D. Zolotarenko).
Author information
Authors and Affiliations
Contributions
Authors A.D. Zolotarenko and S.A. Bruskin formulated the idea of research and developed an experiment, E.V. Chekalin conducted a meta-analysis and construction of graphs of protein-protein interactions, A.D. Zolotarenko assessed the accumulation of transcripts in biopsies. A.D. Zolotarenko and E.V. Chekalin participated in data processing. All authors participated in the writing of the text of the article and in the discussion of the results.
Corresponding author
Ethics declarations
Conflict of interest. The authors declare that they have no conflict of interest.
Statement of compliance with standards of research involving humans as subjects. The study was approved by the Local Ethics Committee at the Institute of General Genetics of the Russian Academy of Sciences and complies with the principles set forth in the Declaration of the Helsinki Agreement of 1964 and its subsequent changes or comparable ethical standards. All patients signed informed voluntary consent forms.
Rights and permissions
About this article
Cite this article
Chekalin, E.V., Zolotarenko, A.D. & Bruskin, S.A. IQGAP Genes in Psoriasis. Russ J Genet 56, 345–353 (2020). https://doi.org/10.1134/S1022795420030047
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795420030047