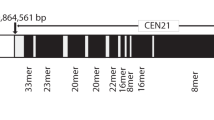
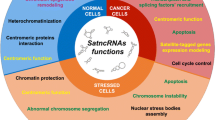
Abstract
Satellite DNA, whose monomers form long arrays of tandem repeat ranging from hundreds or thousands of copies to several million base pairs, makes up at least 10% of the human genome. Application of new methods of sequencing and bioinformatics analysis opens the way to investigate the organization and functioning of human satellite DNA and contributes to the revision of the long-standing view about this part of the genome as “junk DNA.” One of the important features of satellite DNA is its participation in structural rearrangements in the human karyotype. This review examines the mechanisms of participation of satellite DNA in the formation of structural rearrangements, as well as the nature of transcription of tandem repeats in structural rearrangements in the karyotype of normal and tumor cells.
Similar content being viewed by others
REFERENCES
Hemleben, V., Beridze, T.G., Bakhmanz, L., et al., Satellite DNA, Usp. Biol. Khim., 2003, vol. 43, pp. 267—306.
de Koning, A.P., Gu, W., Castoe, T.A., et al., Repetitive elements may comprise over two-thirds of the human genome, PLoS Genet., 2011, vol. 7, no. 12. e1002384. https://doi.org/10.1371/journal.pgen.1002384
Podgornaya, O.I., Ostromyshenskii, D.I., and Enukashvily, N.I., Who needs this junk, or genomic dark matter, Biochemistry (Moscow), 2018, vol. 83, no. 4, pp. 450—466. https://doi.org/10.1134/S0006297918040156
Sullivan, L.L., Chew, K., and Sullivan, B.A., α Satellite DNA variation and function of the human centromere, Nucleus, 2017, vol. 13, pp. 1—9. https://doi.org/10.1080/19491034.2017.1308989
Altemose, N., Miga, K.H., Maggioni, M., et al., Genomic characterization of large heterochromatic gaps in the human genome assembly, PLoS Comput. Biol., 2014, vol. 10, no. 5. e1003628. https://doi.org/10.1371/journal.pcbi.1003628
Liehr, T., Benign and Pathological Chromosomal Imbalances: Microscopic and Submicroscopic Copy Number Variations (CNVs) in Genetics and Counseling, Elsevier, 2014. 220 p.
Alkan, C., Ventura, M., Archidiacono, N., et al., Organization and evolution of primate centromeric DNA from whole-genome shotgun sequence data, PLoS Comput. Biol., 2007, vol. 3, no. 9, pp. 1807—1818. https://doi.org/10.1371/journal.pcbi.0030181
Klein, S.J. and O’Neill, R.J., Transposable elements: genome innovation, chromosome diversity, and centromere conflict, Chromosome Res., 2018, vol. 26, nos. 1—2, pp. 5—23. https://doi.org/10.1007/s10577-017-9569-5
Schueler, M.G. and Sullivan, B.A., Structural and functional dynamics of human centromeric chromatin, Annu. Rev. Genomics Hum. Genet., 2006, vol. 7, pp. 301—313. https://doi.org/10.1146/annurev.genom.7.080505.115613
Peng, J.C. and Karpen, G.H., Epigenetic regulation of heterochromatic DNA stability, Curr. Opin. Genet. Dev., 2010, vol. 18, no. 2, pp. 204—211. https://doi.org/10.1016/j.gde.2008.01.021
Bierhoff, H., Postepska-Igielska, A., and Grummt, I., Noisy silence: non-coding RNA and heterochromatin formation at repetitive elements, Epigenetics, 2014, vol. 9, no. 1, pp. 53—61. https://doi.org/10.4161/epi.26485
Sullivan, B.A. and Karpen, G.H., Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin, Nat. Struct. Mol. Biol., 2004, vol. 11, no. 11, pp. 1076—1083. https://doi.org/10.1038/nsmb845
Hori, T., Shang, W.H., Toyoda, A., et al., Histone H4 Lys 20 monomethylation of the CENP-A nucleosome is essential for kinetochore assembly, Dev. Cell, 2014, vol. 29, no. 6, pp. 740—749. https://doi.org/10.1016/j.devcel.2014.05.001
Bailey, A.O., Panchenko, T., Shabanowitz, J., et al., Identification of the post-translational modifications present in centromeric chromatin, Mol. Cell. Proteom., 2016, vol. 15, no. 3, pp. 918—931. https://doi.org/10.1074/mcp.M115.053710
Black, E.M. and Giunta, S., Repetitive fragile sites: centromere satellite DNA as a source of genome instability in human diseases, Genes (Basel), 2018, vol. 9, no. 12, p. 615. https://doi.org/10.3390/genes9120615
Fournier, A., Florin, A., Lefebvre, C., et al., Genetics and epigenetics of 1q rearrangements in hematological malignancies, Cytogenet. Genome Res., 2007, vol. 118, nos. 2—4, pp. 320—327. https://doi.org/10.1159/000108316
Gardner, R.J.M. and Amor, D.J., Chromosome Abnormalities and Genetic Counseling, Oxford: Oxford Univ. Press, 2018.
Dimitri, P., Corradini, N., Rossi, F., et al., The paradox of functional heterochromatin, Bioessays, 2005, vol. 27, no. 1, pp. 29—41. https://doi.org/10.1002/bies.20158
Scheuermann, M.O., Tajbakhsh, J., Kurz, A., et al., Topology of genes and nontranscribed sequences in human interphase nuclei, Exp. Cell Res., 2004, vol. 301, no. 2, pp. 266—279. https://doi.org/10.1016/j.yexcr.2004.08.031
Giunta, S. and Funabiki, H., Integrity of the human centromere DNA repeats is protected by CENP-A, CENP-C, and CENP-T, Proc. Natl. Acad. Sci. U.S.A., 2017, vol. 114, no. 8, pp. 1928—1933. https://doi.org/10.1073/pnas.1615133114
Amor, D.J., Bentley, K., Ryan, J., et al., Human centromere repositioning “in progress”, Proc. Natl. Acad. Sci. U.S.A., 2004, vol. 101, no. 17, pp. 6542—6547. https://doi.org/10.1073/pnas.0308637101
Jagannathan, M. and Yamashita, Y.M., Function of junk: pericentromeric satellite DNA in chromosome maintenance, Cold Spring Harb. Symp. Quant. Biol., 2017, vol. 82, pp. 319—327. https://doi.org/10.1101/sqb.2017.82.034504
Grewal, S.I. and Jia, S., Heterochromatin revisited, Nat. Rev. Genet., 2007, vol. 8, no. 1, pp. 35—46. https://doi.org/10.1038/nrg2008
Eichler, E.E., Duplication, structure and the evolution of the human genome, Mol. Cytogenet., 2017, vol. 10, no. 1, p. L27.
Metzler-Guillemain, C., Mignon, C., Depetris, D., et al., Bivalent 15 regularly associates with the sex vesicle in normal male meiosis, Chromosome Res., 1999, vol. 7, pp. 369—378.
Crosetto, N., Mitra, A., Silva, M.J., et al., Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing, Nat. Methods, 2013, vol. 10, no. 4, pp. 361—365. https://doi.org/10.1038/nmeth.2408
Tomasetti, C., Li, L., and Vogelstein, B., Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention, Science, 2017, vol. 355, no. 6331, pp. 1330—1334. https://doi.org/10.1126/science.aaf9011
Jackson, S.P. and Bartek, J., The DNA-damage response in human biology and disease, Nature, 2009, vol. 461, no. 7267, pp. 1071—1078. https://doi.org/10.1038/nature08467
Barra, V. and Fachinetti, D., The dark side of centromeres: types, causes and consequences of structural abnormalities implicating centromeric DNA, Nat. Commun., 2018, vol. 9, no. 1, p. 4340. https://doi.org/10.1038/s41467-018-06545-y
Markova, Zh.G., Min’zhenkova, M.V., Musatova, E.V., et al., Unbalanced Y-autosomal translocations without phenotypic manifestations, Med. Genet., 2018, vol. 11, pp. 7—10. https://doi.org/10.25557/2073-7998
Kolomiets, O.L., Lelekova, M.A., Kashintsova, A.A., et al., Detection of disorders of meiosis and spermatogenesis using light, electron and fluorescence microscopy, Androl. Genital.Khir., 2018, vol. 19, no. 1. https://doi.org/10.17650/2070-9781-2018-19-1-00-00
Baranov, V.S. and Kuznetsova, T.V., Tsitogenetika embrional’nogo razvitiya cheloveka (Cytogenetics of Human Embryonic Development), St. Petersburg: N-L, 2007.
Kovaleva, N.V., Homologous Robertsonian translocations: spectrum, sex ratios, and reproductive risks, Russ. J. Genet., 2019, vol. 55, no. 1, pp. 10—23. https://doi.org/10.1134/S1022795419010095
McFarlane, R.J. and Humphrey, T.C., A role for recombination in centromere function, Trends Genet., 2010, vol. 26, no. 5, pp. 209—213. https://doi.org/10.1016/j.tig.2010.02.005
Lee, H., Hayden, K.E., and Willard, H.F., Organization and molecular evolution of CENP-A associated satellite DNA families in a basal primate genome, Genome Biol. Evol., 2011, vol. 3, pp. 1136—1149. https://doi.org/10.1093/gbe/evr083
Arias, E.E. and Walter, J.C., Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells, Genes Dev., 2007, vol. 21, no. 5, pp. 497—518. https://doi.org/10.1101/gad.1508907
Archambault, V., Ikui, A.E., Drapkin, B.J., et al., Disruption of mechanisms that prevent rereplication triggers a DNA damage response, Mol. Cell. Biol., 2005, vol. 25, no. 15, pp. 6707—6721. https://doi.org/10.1128/MCB.25.15.6707-6721.2005
Iwata-Otsubo, A., Dawicki-McKenna, J.M., Akera, T., et al., Expanded satellite repeats amplify a discrete CENP-A nucleosome assembly site on chromosomes that drive in female meiosis, Curr. Biol., 2018, vol. 27, no. 15, pp. 2365—2373. https://doi.org/10.1016/j.cub.2017.06.069
Müller, S. and Almouzni, G., Chromatin dynamics during the cell cycle, Nat. Rev. Genet., 2017, vol. 18, no. 3, pp. 192—208. https://doi.org/10.1038/nrg.2016.157
Choo, K.H.A., Why is the centromere so cold?, Genome Res., 1998, vol. 8, pp. 81—82. https://doi.org/10.1101/gr.8.2.81
Kasinathan, S. and Henikoff, S., Non-B-form DNA is enriched at centromeres, Mol. Biol. Evol., 2018, vol. 35, no. 4, pp. 949—962. https://doi.org/10.1093/molbev/msy010
Aze, A., Sannino, V., Soffientini, P., et al., Centromeric DNA replication reconstitution reveals DNA loops and ATR checkpoint suppression, Nat. Cell Biol., 2016, vol. 18, no. 6, pp. 684—691. https://doi.org/10.1038/ncb3344
Lai, X., Broderick, R., Bergoglio, V., et al., MUS81 nuclease activity is essential for replication stress tolerance and chromosome segregation in BRCA2-deficient cells, Nat. Commun., 2016, vol. 8. 15983. https://doi.org/10.1038/ncomms15983
Jaco, I., Vera, E., and Blasco, M.A., Centromere mitotic recombination in mammalian cells, J. Cell Biol., 2008, vol. 181, no. 6, pp. 885—892. https://doi.org/10.1083/jcb.200803042
Miniou, P., Jeanpierre, M., Blanquet, V., et al., Abnormal methylation pattern in constitutive and facultative (X inactive chromosome) heterochromatin of ICF patients, Hum. Mol. Genet., 1994, vol. 3, no. 12, pp. 2093—2102.
Jeanpierre, M., Turleau, C., Aurias, A., et al., An embryonic-like methylation pattern of classical satellite DNA is observed in ICF syndrome, Hum. Mol. Genet., 1993, vol. 2, no. 6, pp. 731—735.
Hagleitner, M.M., Lankester, A., Maraschio, P., et al., Clinical spectrum of immunodeficiency, centromeric instability and facial dysmorphism (ICF syndrome), J. Med. Genet., 2008, vol. 45, no. 2, pp. 93—99. https://doi.org/10.1136/jmg.2007.053397
Ehrlich, M., The ICF syndrome, a DNA methyltransferase 3B deficiency and immunodeficiency disease, Clin. Immunol., 2003, vol. 109, no. 1, pp. 17—28. https://doi.org/10.1016/S1521-6616(03)00201-8
Wijmenga, C., Hansen, R.S., Gimelli, G., et al., Genetic variation in ICF syndrome: evidence for genetic heterogeneity, Hum. Mutat., 2000, vol. 16, no. 6, pp. 509—517. https://doi.org/10.1002/1098-1004(200012)16:6<509::AID-HUMU8>3.0.CO;2-V
Thijssen, P.E., Ito, Y., Grillo, G., et al., Mutations in CDCA7 and HELLS cause immunodeficiency-centromeric instability—facial anomalies syndrome, Nat. Commun., 2015, vol. 6, p. 7870. https://doi.org/10.1038/ncomms8870
Unoki, M., Funabiki, H., and Velasco, G., CDCA7 and HELLS mutations undermine nonhomologous end joining in centromeric instability syndrome, J. Clin. Invest., 2018, vol. 129, no. 1, pp. 78—92. https://doi.org/10.1172/JCI99751
Mills, R.E., Bennett, E.A., Iskow, R.C., et al., Which transposable elements are active in the human genome?, Trends Genet., 2007, vol. 23, no. 4, pp. 183—191. https://doi.org/10.1016/j.tig.2007.02.006
McNulty, S.M., Sullivan, L.L., and Sullivan, B.A., Human centromeres produce chromosome-specific and array-specific satellite transcripts that are complexed with CENP-A and CENP-C, Dev. Cell, 2017, vol. 42, no. 3, pp. 226—240. https://doi.org/10.1016/j.devcel.2017.07.001
Lemoine, F.J., Degtyareva, N.P., Lobachev, K., et al., Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites, Cell, 2005, vol. 120, no. 5, pp. 587—598. https://doi.org/10.1016/j.cell.2004.12.039
Enukashvily, N.I. and Ponomartsev, N.V., Mammalian satellite DNA: a speaking dumb, Adv. Protein Chem. Struct. Biol., 2013, vol. 90, pp. 31—65. https://doi.org/10.1016/B978-0-12-410523-2.00002-X
Biscotti, M.A., Canapa, A., Forconi, M., et al., Transcription of tandemly repetitive DNA: functional roles, Chromosome Res., 2015, vol. 23, pp. 463—477. https://doi.org/10.1007/s10577-015-9494-4
Trofimova, I. and Krasikova, A., Transcription of highly repetitive tandemly organized DNA in amphibians and birds: a historical overview and modern concepts, RNA Biol., 2016, vol. 13, no. 12, pp. 1246—1257. https://doi.org/10.1080/15476286.2016.1240142
Fournier, A., McLeer-Florin, A., and Lefebvre, C., 1q12 chromosome translocations form aberrant heterochromatic foci associated with changes in nuclear architecture and gene expression in B cell lymphoma, EMBO Mol. Med., 2010, vol. 2, no. 5, pp. 159—171. https://doi.org/10.1002/emmm.201000067
Alitalo, T., Tiihonen, J., Hakola, P., et al., Molecular characterization of Y;15 translocation segregating in a family, Hum. Genet., 1988, vol. 79, no. 1, pp. 29—35. https://doi.org/10.1007/bf00291705
Ferreira, D., Meles, S., Escudeiro, A., et al., Satellite non-coding RNAs: the emerging players in cells, cellular pathways and cancer, Chromosome Res., 2015, vol. 23, no. 3, pp. 479—493. https://doi.org/10.1007/s10577-015-9482-8
Ting, D.T., Lipson, D., Paul, S., et al., Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers, Science, 2011, vol. 331, no. 6017, pp. 593—596. https://doi.org/10.1126/science.1200801
Eymery, A., Horard, B., Atifi-Borel, M.El., et al., A transcriptomic analysis of human centromeric and pericentric sequences in normal and tumor cells, Nucleic Acids Res., 2009, vol. 37, no. 19, pp. 6340—6354. https://doi.org/10.1093/nar/gkp639
Hall, L.L., Byron, M., Carone, D.M., et al., Demethylated HSATII DNA and HSATII RNA foci sequester PRC1 and MeCP2 into cancer-specific nuclear bodies, Cell Rep., 2017, vol. 18, no. 12, pp. 2943—2956. https://doi.org/10.1016/j.celrep.2017.02.072
Bersani, F., Lee, E., Kharchenko, P.V., et al., Pericentromeric satellite repeat expansions through RNA-derived DNA intermediates in cancer, Proc. Natl. Acad. Sci. U.S.A., 2015, vol. 112, no. 49, pp. 15148—15153. https://doi.org/10.1073/pnas.1518008112
Eymery, A., Callanan, M., and Vourc’h, C., The secret message of heterochromatin: new insights into the mechanisms and function of centromeric and pericentric repeat sequence transcription, Int. J. Dev. Biol., 2009, vol. 53, nos. 2—3, pp. 259—268. https://doi.org/10.1387/ijdb.082673ae
Alexiadis, V., Ballestas, M.E., Sanchez, C., et al., RNAPol-ChIP analysis of transcription from FSHD-linked tandem repeats and satellite DNA, Biochim. Biophys. Acta, 2007, vol. 1769, no. 1, pp. 29—40. https://doi.org/10.1016/j.bbaexp.2006.11.006
Hoshi, N., Fujita, M., Mikuni, M., et al., Seminoma in a postmenopausal woman with a Y;15 translocation in peripheral blood lymphocytes and a t(Y;15)/45,X Turner mosaic pattern in skin fibroblasts, J. Med. Genet., 1998, vol. 35, no. 10, pp. 852—856. https://doi.org/10.1136/jmg.35.10.852
Gravholt, C.H., Fedder, J., Naeraa, R.W., et al., Occurrence of gonadoblastoma in females with Turner syndrome and Y chromosome material: a population study, J. Clin. Endocrinol. Metab., 2000, vol. 85, no. 9, pp. 3199—3202. https://doi.org/10.1210/jcem.85.9.6800
Jehan, Z., Vallinayagam, S., Tiwari, S., et al., Novel noncoding RNA from human Y distal heterochromatic block (Yq12) generates testis-specific chimeric CDC2L2, Genome Res., 2007, vol. 17, no. 4, pp. 433—440. https://doi.org/10.1101/gr.5155706
Kuznetsova, T.V., Enukashvili, N.I., Trofimova, I.L., et al., Localization and transcription of the centromeric heterochromatin of chromosome 1 in human embryonic and extraembryonic tissues, Med. Genet., 2012, vol. 11, no. 4(118), pp. 19—24.
Trofimova, I.L., Enukashvili, N.I., Kuznetsova, T.V., et al., Transcription of satellite DNA in human embryogenesis: a review of the literature and our own data, Med. Genet., 2018, vol. 17, no. 3, pp. 3—7. https://doi.org/10.25557/2073-7998.2018.03.3-7
Collins, F.S., Green, E.D., and Guttmacher, A.E., A vision for the future of genomics research, Nature, 2003, vol. 422, no. 6934, pp. 835—847. https://doi.org/10.1038/nature01626
Miga, K.H., Completing the human genome: the progress and challenge of satellite DNA assembly, Chromosome Res., 2015, vol. 23, no. 3, pp. 421—426. https://doi.org/10.1007/s10577-015-9488-2
Miga, K.H., Newton, Y., Jain, M., et al., Centromere reference models for human chromosomes X and Y satellite arrays, Genome Res., 2014, vol. 24, no. 4, pp. 697—707. https://doi.org/10.1101/gr.159624.113
Jain, M., Koren, S., Miga, K.H., et al., Nanopore sequencing and assembly of a human genome with ultra-long reads, Nat. Biotechnol., 2018, vol. 36, no. 4, pp. 338—345. https://doi.org/10.1038/nbt.4060
Funding
This work was financially supported by the Russian Foundation for Basic Research (no. 18-34-00279).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by K. Lazarev
Rights and permissions
About this article
Cite this article
Puppo, I.L., Saifitdinova, A.F. & Tonyan, Z.N. The Role of Satellite DNA in Causing Structural Rearrangements in Human Karyotype. Russ J Genet 56, 41–47 (2020). https://doi.org/10.1134/S1022795419080155
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795419080155