Abstract
This review deals with important issues in regulation of terminal alternative oxidase (AOX) of plant mitochondria, which are currently investigated and discussed. AOX ensures electron transport along the alternative nonphosphorylating cyanide-resistant pathway in mitochondrial ETC and plays an important role in the maintenance of redox balance in the cell and defense against stress. Principal results concerning AOX signaling were obtained in the works dealing with expression of the АОХ1а gene of Arabidopsis thaliana (AtAOX1a). Reports about АОХ expression in different plant species were analyzed. Mechanisms of transcriptional and posttranscriptional control over expression of АОХ genes were described. The role of some cis-regulatory elements of AtАОХ1а promoter and related transcription factors was considered. It was shown that transduction of the signal causing expression of АОХ genes may follow anterograde and retrograde pathways. Information about mitochondrial retrograde regulation of AOX and ROS-dependent and ROS-independent signal pathways of induction of АОХ expression is given. A hypothetic sequence of signal events of light induction of АОХ1а expression in wheat leaf cell in the course of greening was worked out. Interactions between AOX signaling and hormonal signal pathways, as well as induction of АОХ by the signals of plastid origin, were described. In conclusion, it was highlighted that AOX is integrated into the common signal pathway of the cell and acts as a target and a regulator of signal events.
Similar content being viewed by others
INTRODUCTION
Along with the main (cytochrome) pathway, the inner membrane of plant mitochondria houses an alternative pathway (AP) via terminal oxidase called alternative oxidase (AOX). AOX that catalyzes four-electron reduction of oxygen by ubiquinone to water is resistant to cyanide. AP of electron transport bypasses two points of generation of an electrochemical proton gradient (complexes III and IV) and the energy of ubiquinone oxidation by oxygen is released as heat. This circumstance reduces energy efficiency of respiration but removes respiratory control that regulates the rate of ATP and NADH production depending on cell needs [1]. AP was discovered not only in higher plants but also in the representatives of all the kingdoms (except for Archaebacteria): bacteria, algae, moss, ferns, some fungi, and even in slow-moving or stationary animals (sponge, actinia, nematode, mollusk, etc.) [1].
The original interest in AOX was aroused by its participation in thermogenesis of floral tissues in some representatives of the Araceae family. The rise in temperature of thermogenic tissues promotes volatilization of substances that attract pollinator insects. This phenomenon was described by Lamarck way back in the 18th century, and cyanide-resistant respiration was discovered in the 1930s [2]. The nonphosphorylating nature of AP brought up questions for a long time concerning the role of AP in nonthermogenic tissues. The nature, mechanisms of regulation, and physiological significance of AOX are dealt with in numerous reviews [1, 3–7]. At present, the majority of researchers share an opinion that, at the cellular and biochemical levels, operation of AP contributes to the maintenance of redox equilibrium in mitochondrial ETC owing to quicker oxidation of NADH and prevents generation of excess ROS. This affects metabolism not only in mitochondria but also in the whole cell. It is generally accepted that AOX ensures metabolic plasticity of the cell and plant under changing environmental conditions [6].
The alternative pathway of respiration is not only a basic issue. Alternative oxidase was proposed as a functional marker of cell reprogramming under stress conditions for breeding of highly productive and resistant genotypes [8], for determination of plant response to changes in environmental conditions (global warming and elevation of СО2 in the atmosphere) [9], and for control over mitochondrial diseases in humans [10]. Owing to the application of modern methods of physical chemistry and molecular genetics, considerable progress has been made in the last two decades in the investigation of structure of proteins and genes as well as AOX functions. AOX is believed to act not only as a target but also as a regulator of stress signal [11]. At the same time, the investigation of certain aspects of AOX signaling at the molecular level is only beginning [1, 7, 12, 13].
This review deals with the available data concerning AOX genes, signal pathways and mechanisms of induction of their expression, and integration of AOX signaling with other signal pathways in the cell; the role of AOX in the maintenance of signal intracellular interactions was analyzed; the mechanisms of posttranslational modifications of AOX protein were discussed. The review also covers our own experimental data indicating that AOX participates in modulation of mitochondrial function.
AOX GENES AND POLYMORPHISM IN AOX SEQUENCES
In dicotyledons, AOX is encoded by two subfamilies of nuclear genes, АОХ1 and АОХ2, while it is by subfamily АОХ1 alone [14] in monocotyledons, except for some representatives of an ancient but early degraded group from the family Araceae [15]. The formation of two subfamilies and variation in the number of genes apparently result from duplication and divergence [14, 15]. АОХ2 is present in algae and gymnosperms but was not found in mosses, ferns, and lycopods. In the fungal genome, АОХ gene of only one subfamily was found. Arabidopsis thaliana contains four АОХ1 genes (a–d) and one АОХ2 gene, while soybean Glycine max, on the contrary, contains one АОХ1 gene and two АОХ2 genes (а, b) [14]. Rice has four АОХ1 genes (a–d) [14]; in wheat, only genes АОХ1a and АОХ1c [16] were identified, although the discovery of other genes from this subfamily (AOX1d, AOX1e) has not been ruled out [17]. The genome of carrot Daucus carota L. is unique, it contains two genes from each subfamily: AOX1 (a, b) and AOX2 (a, b) [18].
Most of the genes have a conserved intron-exon structure composed of four exons alternating with three introns. The variation of АОХ genes’ structure among the species is associated with a deletion or insertion of an intron [14]. The investigation of АОХ induction at the level of transcription was conducted for the most part in plants with a sequenced genome: Arabidopsis thaliana (L.) Heynh., A. lyrata L., Nicotiana tabacum L., Zea mays L., and some other species; this makes it possible to determine molecular mechanisms of signaling, regulation of AOX activity, and its role in stress response.
АОХ genes may show single nucleotide polymorphism (SNP) usually associated with single substitutions. In АОХ, SNP may occur both in exons and introns [14]. SNP was detected in several regions of gene АОХ2 in three cultivars of Olea europaea L. [19]. Polymorphous sites of АОХ with an insertion/deletion and copying DNA fragments were also found. In particular, polymorphism of АОХ in carrot (DcAOX2a) was revealed in intron 3 [20]. The functional role of all the found SNPs in АОХ is not clear. It is known that the change from Lys (N) to Asn (K) at amino acid position 96 in AOX protein from watermelon resulted from SNP in a single watermelon AOX gene: ClAOX [21]. It was shown that this SNP is associated with quantitative trait locis responsible for resistance to hypothermia [22]. Variations in gene AOX sequence were proposed as molecular markers for stress resistance of a species (including plant breeding) and genetic variation accounting for individual differences between the plants of the same species [8, 14]. As a result of point mutation in rice gene АОХ1а, a single nucleotide replacement was produced; substitution of Lys(71) for Asn(71) in the protein improved the tolerance of the rice variety to low temperatures [23].
Another aspect of AOX genes’ polymorphism is associated with microheterogeneity of the 3'-untranslated region (3'-UTR) of АОХ transcripts as a result of alternative polyadenylation [14]. Modulation of 3'‑UTR length may considerably affect the regulation of AOX expression, although appropriate indications are not numerous. In the 3'-UTR of maize АОХ1а, a putative site of binding with microRNA (miR163) was identified [14, 24]. This site is located between two cleavage sites, which suggests that only transcripts with long enough 3'-UTR may undergo microRNA silencing. In maize seedlings, approximately half of mRNA molecules of AOX1a whose transcription was induced by treatment with H2O2 had short 3'-UTR without the site of microRNA binding; therefore, they escaped microRNA silencing [24]. In Daucus carota, AОХ2a bore a repetitive deletion in intron 3, which is supposed to affect the site of binding with microRNA [20]. Numerous variations of 3'-UTR length in АОХ2 were detected in different cultivars of an olive-tree (Olea europaea L.) [19]. It may be a conserved trait in cultivars with different ability of adventitious rooting associated with stress-induced expression of АОХ2 [19].
Although information about polymorphism of AOX genes is still incomplete, it becomes evident that polymorphhism of АОХ is an evolutionary mechanism regulating АОХ expression in response to different signals, which improves plant tolerance to stress.
TRANSCRIPTIONAL AND POSTTRANSCRIPTIONAL CONTROL OVER EXPRESSION OF AOX GENES
Regulation of gene transcription activity greatly depends on the structure of its promoter region. The most thoroughly investigated are promoters of AOX genes in A. thaliana [25, 26] and Glycine max [27]. Gene АОХ1а from A. thaliana (AtAOX1a) serves as a model for investigation of transcriptional control over the expression of АОХ under stress [5, 26, 28].
Promoter region comprises cis-acting regulatory elements (CAREs) that, as a rule, are located close to TSS (transcription start site). These elements can interact with RNA polymerase and transcription factors (TF) that govern gene expression.
Analysis of the AtAOX1a promoter by means of deletions and mutagenesis revealed a 93-bp-long region (a segment between the 313th and 406th nucleotide bases upstream of TSS; hereinafter, all the positions upstream from TSS are given with the “–” sign) designated as regulator of expression of AtAOX1a involved in the mitochondrial retrograde pathway [13, 28]. This region of promoter regulates expression of the gene upon exposure to antimycin, an inhibitor of ETC complex III, and monofluoroacetate (MFA), an inhibitor of TCA cycle (aconitase). Subsequently, a CARE was discovered and designated as MDM (mitochondrial dysfunction motif) with a CTTGNNNNNCA(A/C)G sequence, which regulates expression of 24 so-called MDS (mitochondrial dysfunction stimulon) genes, including АОХ1а [29].
Analysis of AtAOX1a 1800-bp-long promoter upstream from TSS revealed 15 CAREs, out of which 12 were functionally active (A1, А2, B2, C, D, E, F, G, H, I1, I2, and J) and participated in regulation of the gene transcription in response to exposure to stress agents, such as Н2О2 and rotenone (an inhibitor of ETC complex I) or upon their joint action [26]. Under normal conditions, the AtAOX1a promoter is repressed under the control of element B2 (CGTGAG, –1576 bp). This element is the site of binding with TF ABI4 (ABA-insensitive 4). ABI4 is believed to be a repressor of mitochondrial retrograde regulation (MRR) since abi4 mutants showed an elevated content of the gene transcripts [30].
CAREs A2, E, I1, and I2 operate as activators of MRR. Upon exposure to stress agents, repression decreased and other CAREs showed a positive regulatory activity, which points to a combinative regulation of gene expression [26].
Elements B and I bear site ABRE (ABA-responsive element) (ACGT), and element I1 (ACGTG, –93 bp) is located in immediate proximity to I2 producing an ABRE-ABRE pair. ABRE is recognized by TF bZIP (leucine zipper); two copies of ABRE probably ensure a high sensitivity of the gene to ABA [26].
CARE I1 in the AtАОХ1а promoter is also overlapping with a light-sensitive element of G-box (CACGTG), which points to the existence of direct mechanisms connecting gene expression with photosynthetic function [26, 31, 32]. Reports about photosensitive elements of the АОХ promoter and light-dependent nature of АОХ induction are given below in the section dealing with signal pathways.
Genetic screening revealed sites (T/G)CGTGT and W-box ((a/t)TTGAC(a/c/t)) in the AtAOX1a promoter, which bind TF from NAC and WRKY families, respectively [28, 33]. In A. thaliana, ANAC017, ANAC013, and other ANAC are positive regulators of AtAOX1a expression [29, 34], whereas WRKY are both positive and negative regulators of AtAOX1a expression [33].
Analysis of 1000-bp-long promoter of another AOX gene, AtAOX1c, that is not induced in response to oxidative stress showed nine CAREs (A–I) predominantly located in the region of 500 bp upstream from TSS [25]. Out of them, seven elements were also present in the promoter region of soybean AOX gene GmAOX2b, which is not stress-inducible [27]. Deletions of large fragments and CAREs of AtAOX1c and GmAOX2b promoters have shown that the elements operated in different ways displaying both negative and positive regulation of gene expression depending on the examined tissue and tested signals. For instance, all the elements of AtAOX1c promoter except C (–342 bp) in line ΔAt2g19080 (with elevated content of transcripts of AOX1c and AOX protein) showed a positive regulation of expression, whereas some elements, specifically E (–230 bp), operated as negative regulators in cell suspension and in the leaves of A. thaliana plants, ecotype Col-0 [25]. Genome-wide test based on Expression Angler data analysis tool and ACT (arabidopsis coexpression tool) revealed common functional elements in promoters of AtAOX1c and the genes whose expression is associated with the processes of cell division and growth. This suggested that the expression of AtAOX1c is regulated by the signals related to growth and development [25].
We have analyzed promoters of all the AtAOX genes and showed common CAREs involved in formation of sensitivity to various stresses in all the genes [35, 36]. G-box (CACGTG) is present in promoters of not only АОХ1а but also of АОХ2 [36]. Promoters of genes AOX1a, AOX1c, and AOX1d are enriched in such CAREs as MYB4-BSM (binding site motif), SORLEP3, SORLIP2, and CCA1-BSM, which points to a dependence of their expression on changes in operation of chlETC (chloroplast ETC) in the light. In A. thaliana, CCA1-BSM may participate in phytochrome-regulated expression of genes LHCB encoding proteins binding chlorophyll a/b [37]. A complete sequence of UVBoxANAC13 (CCAAGG) responsible for a high sensitivity to UV-B [38] was detected in genes AOX1a and AOX1c [35]. It is interesting that promoters of all АОХ1 genes contained a reduced version of this element (CCAAG) characteristic of the genes with a deferred (as compared with the genes bearing a complete UVBoxANAC13 sequence) UV-B-induced expression [38]. Light-sensitive CAREs called MRECHS-like box and ACECHS-like box were found in promoters of all AtАОХ genes except for AOX1b [35]; they are abundant in promoters of UV-induced genes such as CHS (gene of chalcone synthase) [38]. The presence of common CAREs points to a complementary interaction between AtАОХ genes at a transcription level within a certain range of stress response. For instance, upon exposure to elevated physiological dose of UV-B and moderate illuminance (400 µmol/(m2 s)), A. thaliana plants antisense by AOX1a showed a compensatory induction of other AOX genes’ expression [35, 36]. At the same time, the activity of AOX genes upon exposure to bright light was probably regulated via different signal pathways. This was suggested by the results of cluster analysis that made it possible to attribute AOX genes to different clusters depending on the character of their expression [36].
Mechanisms of posttranscriptional control over the expression of AOX genes associated with RNA processing are poorly investigated. Pioneering data concerning the regulation of AOX genes at a posttranscriptional level were obtained in the work dealing with SgАОХ expression in the spadix of Sauromatum guttatum [39]. A multiple intensification of gene expression was detected, whereas the rate of transcription in isolated nuclei did not change. It is believed that, in addition to polyadenylation detected in several AOX genes [14], processing of АОХ RNA greatly depends on capping associated with phosphorylation of the 5'-end of transcript and attachment via a triphosphate bridge of 7-methylguanosine (cap structures) [40]. In the cells of animals, capping is partially regulated by the target of rapamycin (TOR): a serine-threonine protein kinase. TOR is present in all the eukaryotes. Within multimolecular signal complexes (animals and yeast have two of them and plants have only one), TOR regulates cell growth and survival activating rapid synthesis of protein, which is especially important under stress. In A. thaliana plants, TOR may be an acceptor of protein kinase KIN10, probably via an intermediary in the nucleus: cyclin-dependent kinase Е1 (CDKE1) that induces expression of AOX1a [40].
EXPRESSION PATTERNS AND TRIGGERS OF АОХ ACTIVATION
Active investigation of AOX genes’ expression started after cloning the first cDNA of АОХ from Sauromatum guttatum [41]. To date, much information has been obtained about the pattern of АОХ expression in different species. It was shown that the expression of АОХ may be constitutive in the course of normal growth and development, whereas it may considerably change under stress conditions [5, 14, 42]. Orthologous genes usually show the same pattern of expression; Aox2a and Aox2b from soybean and cowpea may exemplify this situation [43]. However, opposite cases exist. Orthologous genes АОХ1а and АОХ1с of wheat and rice showed different patterns of expression in the course of leaf greening [44, 45].
Expression of АОХ was induced in response to abiotic and biotic factors: temperature [46, 47], salinity [48], drought [49], hypoxia [50], high illuminance [31, 36, 51, 52], UV-B radiation [35], heavy metals [52, 54], ozone [55], nitrogen deficiency [56], pathogens [57], and other stress agents [52, 58]. Some literature reports about the expression of АОХ in higher plants are summarized in the supplementary data (Supplementary Table S1); this table shows that the expression of AOX genes differs and expression patterns of the same AOX gene may depend on the type of tissue, growing conditions, and duration of exposure to a stress agent.
Genes from subfamily АОХ1 are usually stress-induced. АОХ1а of A. thaliana is most sensitive to stress but this gene does not necessarily show such a response in other plant species (Supplementary Table S1). The expression of АОХ2 is often regulated by the processes of growth and development, which does not rule out the participation of AOX2 in response to stress agents. In particular, the expression of all four genes of both AOX subfamilies in Daucus carota was organ- and growth-specific [18]. Gene DcAOX1a showed a rather constitutive expression in the leaves and in the course of de novo formation of secondary phloem in the roots of explants. Induction of AOX genes may be triggered by mitochondrial dysfunctions caused by the effect of ETC inhibitors [28, 51] and by mutations of the genes encoding mitochondrial proteins of ETC complexes [59, 60]. It is believed that defects of ETC caused by gene mutations and inhibitors of ETC and TCA cycle generate redox signals that are transduced to the nucleus and initiate expression of АОХ and other (not only respiratory) genes [26, 28, 60]. Expression of AOX (by the example of AtAOX1a) is a model widely used for investigation of mitochondrial retrograde signaling.
Thus, differential expression of AOX genes is associated with genotypic and phenotypic diversity of the cells, regulation of gene activity by arriving signals, and control over АОХ expression at different steps of realization of genetic information. This ensures a plasticity of plant cell metabolism, which involves the energy dissipating function of mitochondria [5, 8, 14, 34].
SIGNAL PATHWAYS OF REGULATION OF АОХ EXPRESSION
Transduction of a signal causing expression of АОХ may follow anterograde or retrograde pathways [11, 13, 30]. Anterograde regulation of gene expression implies a descending (top-down) pathway from the nucleus to the organelle. Retrograde regulation follows an ascending (bottom-up) pathway from the organelles to the nucleus [12, 61]. Anterograde signaling is believed to proceed at several levels from transcriptional to posttranslational; as a rule, the signals are associated with the processes of plant growth and development and exposure to external factors. Retrograde regulation of gene expression occurs in response to changes in internal conditions and enables organelles to affect the expression of nuclear genes [12, 30, 61].
Mitochondrial retrograde regulation of АОХ expression. Mitochondrial retrograde regulation (MRR pathway) was described in detail; its existence is confirmed not only by the data about induction of АОХ expression by inhibitors of ETC and TCA cycle and by defects of ETC complexes but also by the presence of the MRR region in АОХ promoter (by the example of AtAOX1a) necessary for the induction of gene expression [28, 34]. Inhibitors (antimycin and monofluoroacetate) imitate, respectively, ROS-dependent and ROS-independent signal pathways of induction of AOX genes’ expression [11, 13, 28].
Dysfunctions of ETC caused by a disturbance of electron flow lead to the production of excess ROS: superoxide radical (\({\text{O}}_{2}^{{\centerdot - }}\)) and hydrogen peroxide (Н2О2). \({\text{O}}_{2}^{{\centerdot - }}\) is generated in complexes I, II, and III; electrons are donated to oxygen by а ubisemiquinone intermediate [13]. \({\text{O}}_{2}^{{\centerdot - }}\) produced within ETC is unstable (its lifetime is 10–12 s) and quickly neutralized to Н2О2 with the participation of Mn-dependent superoxide dismutase (MnSOD) or spontaneously. It was shown that overexpression of MnSOD reduced expression of АОХ1а and АОХ1b in rice plants exposed to cold stress [62], and MnSOD knockout line of Arabidopsis thaliana showed more intense expression of AOX1а in response to treatment with AlCl3 as compared with the wild type line [63]. These data indicate that the content of ROS in mitochondria closely related to the activity of MnSOD may be a primary signal causing MRR-dependent induction of АОХ expression. Н2О2 is a secondary messenger of ROS-dependent MRR expression of АОХ [13].
It was believed earlier that H2O2 induces expression of AOX by means of direct oxidation or modulation of TF phosphorylation controlling the initiation of transcription [64]. However, it became known that antimycin (an inhibitor of complex III) caused a quicker induction of АОХ1 expression in tobacco cells than exogenous hydrogen peroxide accumulating in intracellular space [65]. This suggested that ROS of mitochondrial origin more efficiently induce expression of АОХ1 than cytoplasmic or chloroplastic ROS. In this work, it was also shown that bongkrekic acid (an inhibitor of mitochondrial permeability transition pore (PTP) involved in triggering apoptosis in animals) blocked antimycin-, \({{{\text{H}}}_{2}}{\text{O}}_{2}^{ - }\), and SA-dependent induction of АОХ1. This gave rise to an alternative hypothesis accounting for ROS-dependent induction of АОХ expression worked out by analogy with triggering the process of programmed cell death (PCD) in animals [65, 66]. ROS (and in particular hydrogen peroxide) accumulating under stress exposure in intracellular space and able to easily cross cell membranes may cause mitochondrial dysfunctions and opening of permeability transition pores (PTP), which is accompanied by the dissipation of membrane potential and is a crucial stage in АОХ induction signaling [65, 66]. Temporary opening of PTP is believed to be an important mechanism of cellular redox signaling and a mode of communication between mitochondria and the nucleus [65].
NAC domain transcription factors (ANAC017 and ANAC013) are direct activators of АОХ1а transcription [29, 34] (see Fig. 1). ANAC017 is a constitutively expressed TF bound with endoplasmic reticulum (ER) via the C-terminal transmembrane domain. In response to antimycin or upon mitochondrial dysfunctions, the C-terminal domain is detached by rhomboid protease and N-terminal domain is released from ER, migrates to the nucleus, and binds to the AOX1а promoter. A rhomboid protease cleavage site is present in ANAC017 between the two domains, and inhibitors of rhomboid protease reduce antimycin-induced expression of AOX1а [34]. Rhomboid protease is assumed to be activated by H2O2 of mitochondrial origin as ANAC017 mutants usually do not express АОХ1а upon treatment with exogenous H2O2. ANAC017 binds directly with the (T/G)CGTGT site of the AtAOX1a promoter and can activate other TF from the NAC family (for instance, ANAC013) that are positive regulators of АОХ1а expression. However, other NAC transcription factors cannot compensate for the absence of ANAC017 [29].
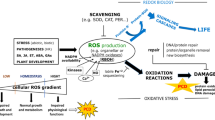
Signal pathways of regulation of AОХ genes’ expression. External and internal (mitochondrial and plastidal) signals transmitted to the nucleus for activation of АОХ (based on the diagrams given in [1, 5, 12]). Pathway of mitochondrial retrograde regulation (MRR pathway) of АОХ expression (by the example of AOX1a from Arabidopsis thaliana, AtAOX1a) comprises arrival of a signal from mitochondria (ROS, citrate, or redox signal) with subsequent release of a transcription factor (TF) of the NAC family (NAC17) from endoplasmic reticulum (ER), which (the same as other NAC TFs) is a positive regulator of AtAOX1a expression. ABI4 is a repressor of AtAOX1a. WRKY TFs may be positive (WRKY63) or negative (WRKY15/40) regulators of АОХ1а expression. Another pathway of signal transduction involves activation of protein kinase (KIN10) that interacts in the nucleus with a mediator complex CDKE1 inducing АОХ1а. Auxin and TF MYB29 are negative regulators of АОХ1а expression.
The promoter of AtАОХ1a is controlled by different TFs that directly interact with regulatory elements and affect the expression of AtAOX1a [5]. It was detected that WRKY are bound with three W-box sites (–291, ‒373, and –412 bp) of the AtAOX1a promoter. WRKY15 and WRKY40 are repressors of retrograde expression of АОХ1а and two other nucleus-encoded genes of mitochondrial proteins of A. thaliana: NDB2 (gene of “external” NADPH dehydrogenase) and UPOX (UP-regulated by OXidative stress) sensitive to oxidative stress [33]. While ABI4 is a repressor of АОХ1а under normal conditions, WRKY15 and WRKY40 restrict the expression of АОХ1а upon ETC dysfunction, specifically, after antimycin treatment. Presumably, WRKY63 operates as an activator of АОХ1а expression. It was found that WRKY63 participates in plant response to high illuminance [33]. The researchers believe that WRKY40 and WRKY63 compete for the binding sites of the gene promoter. It was shown that auxin and TF MYB29 are negative regulators of AtАОХ1а expression operating via hormonal signal pathways and interacting with ERFs (ethylene response factors) and WRKY transcription factors [30, 67].
Signal transduction from mitochondria may also proceed via the activation of KIN10 protein kinase interacting with CDKЕ1, which is a component of the nuclear intermediary complex inducing АОХ1а [40]. In rao1 mutants, it was shown that CDKE1 participates in the perception of any cell signals, including those of anterograde origin (for instance, caused by accumulation of H2O2 or chilling) with subsequent induction of АОХ1а.
Subdivision of АОХ MRR signaling into ROS-dependent and ROS-independent pathways is relative since ROS are potential molecules of transduction of signals received from mitochondria owing to inhibition by both antimycin and MFA [28]. The generation of ROS was detected in tobacco cell suspension exposed to 5 mM MFA [68]. However, this does not rule out a possibility of signal transduction to the promoter region [13, 28]. This is suggested by the fact that different concentrations of MFA regulating expression of АОХ may differently affect ROS formation until the lacking effect [28]. It was also shown that WRKY transcription factor participates in antimycin- and MFA-dependent AtAOX1a induction via MRR pathway; however, mutants in the region of W-box more intensely responded by АОХ1а expression to antimycin than to MFA.
In addition to Н2О2, there are other signal molecules participating in the regulation of АОХ expression: NO, salicylic acid (SA), jasmonic acid, ethylene, and Ca2+ ions [12, 13]. It is well known that plants accumulate Н2О2, NO, and SA in the course of development of hypersensitivity reaction (HSR) in response to a pathogen attack; in addition, the expression of АОХ1а becomes more intense and the content of AOX protein rises [57]. The development of HSR is a good model for the investigation of signal pathways; however, the stages of these pathways have not been decoded. It was shown that the generation of superoxide explosion (“\({\text{O}}_{2}^{{\centerdot - }}\) burst”) in response to inoculation of Nicotiana tabacum with pathogenic bacteria depended on the presence of АОХ1а in the plant genome [57]. It was the generation of \({\text{O}}_{2}^{{\centerdot - }}\) in mitochondria that initiated HSR, although other ROS also participated in its development. The absence of АОХ1а delayed \({\text{O}}_{2}^{{\centerdot - }}\) burst, brought about colocalization of NO and ONOO– (peroxynitrite: a product of interaction between \({\text{O}}_{2}^{{\centerdot - }}\) and NO), and development of HSR in response to a pathogenic attack, the same as to antimycin inhibiting complex III. However, an intense superoxide burst occurred later (as compared with wild type). ONOO– participates in the nitration of tyrosine residua of protein, which affects their modification, sensitivity to proteases, and development of PCD. On the contrary, the overexpression of АОХ1а prevented a superoxide burst induced by antimycin. It was supposed that electron transport (evidently, at the level of complex III) is a target for a reaction generating a signal to HSR development, and AOX is a key determinant of mitochondrial dynamics of ROS in response to pathogenic attack [57, 66]. In early stages of HSR, the reason for superoxide burst is a coordinated joint suppression of gene expression and activity of AOX and MnSOD.
Intensification of АОХ1а expression as a defense response to biotic stress is most probably associated with the effect of SA. At low concentrations (lower than 1 mM), SA interacts as a quinone analog with complexes I and II impairing electron transport in ETC, which triggers signal pathways leading to expression of AOX genes [66]. At the same time, SA initiates the expression of АОХ not directly via TFs that bind with the gene promoter (SA-sensitive element as1 is absent from promoter AtАОХ1а) but more likely at a posttranscriptional level [26]. It is believed that EDS1 (enhanced disease susceptibility 1) and PAD4 (phytoalexin deficiency 4)—lipase-like proteins controlling HSR response, accumulation of SA, and SA-dependent signaling but operating in the upstream zone of SA signaling—regulate expression of АОХ1а (as was shown for AtAOX1a). It is possible that the EDS1/PAD4 signal pathway is unique to induction of АОХ1а expression; this accounts for the lack of coexpression of АОХ1а and NDB2 observed in some cases upon exposure to Н2О2 or rotenone–triggers of MRR pathway [26].
Proteins EDS1 and PAD4 are involved as intermediaries in transduction of the PCD signal stipulated by activation of Toll-like receptors of immune response [69]. AOX is known to participate in the PCD process. The mitochondrial pathway of signaling involved in the majority of apoptosis forms is notable for a sharp rise in ROS preceding the formation of PTP and release of cytochrome c and endonucleases to intermembrane space of mitochondria and then to cytoplasm. In animals, the release of cytochrome с is a key event triggering a cascade of reactions designed to activate caspases and realization of PCD. Regulation of the mitochondrial pathway of PCD involves mechanisms controlling the level of ROS. It was shown that electron transfer along AP prevents the accumulation of ROS and maintains mitochondrial respiration when CP is restricted by inhibitors or compounds inducing PCD [69]. Thus, AOX regulates the cellular level of ROS, which, in turn, determines the susceptibility to development of apoptosis; therefore, AOX was called a survival protein [69].
The role of Ca2+ in MRR-dependent induction of АОХ1а is less clear. The accumulation of Ca2+ in the cell is a sign of developing stress. In animal cells, mitochondria are a depositary of Ca2+ ions and they exchange them with ER-producing calcium microdomains. In plant cells, calcium exchange between compartments is most likely performed by conserved carrier proteins, in particular, by Ca2+/H+-antiporters from the LETM (Leucine zipper, EF hand-containing Transmembrane Protein) family located in mitochondria [11]. In A. thaliana plants exposed to salt stress, Ca2+ exchange between mitochondria and ER was shown to depend on the induction of АОХ1а. In this case, Н2О2-stimulated synthesis of TF WRKY15 suggested that MRR participated in stress response with induction of АОХ1а playing an important role [70]. It is known that ER sends signals to the nucleus in the course of induction of response to protein misfolding (UPR, Unfolded Protein Response), which is an evolutionary conserved transcriptional response necessary for the preservation of ER homeostasis upon accumulation of unfolded proteins. UPR may participate in Са2+-mediated induction of AOX1a regulating activity of the calcium pump in ER, which points to a close relationship between ER, nucleus, and MRR [70].
Anterograde regulation of expression of АОХ genes. Transduction of a signal causing expression of АОХ along the anterograde pathway may be triggered by ROS or other signal molecules of intracellular origin and external agents of different nature [11–13]. It is believed that expression of genes of mitochondrial proteins encoded in the nucleus to a great extent represents anterograde control over the function of mitochondria [12]. Intensification of expression of the genes encoding mitochondrial proteins was detected upon seed germination and in the course of progression through other important phases of plant development. This points to a coordination between gene expression and processes of growth and development and to their involvement in hormonal signaling of the whole plant [12].
Expression of AOX genes in A. thaliana was mainly detected in the organs with a potentially high rate of metabolism [42]. Intense expression of AtAОХ2 was detected in the seeds, and that of AtAОХ1b was in young inflorescences; the expression of AtAOX1d was functionally timed to senescence. Expression of all AtАОХ genes also depended on the phase of growth and development [42]. Close to TSS, the promoter of gene AtАОХ1с bears elements of site II [25] interacting with TF from the TCP family (Teosinte branched 1 Cycloidea Proliferating cell factor), class II—negative factors of cell division [67]. TCP, class I, are considered to be positive regulators of cell growth and proliferation. However, specificity of sequence of the site binding TCP of both classes is not absolutely conserved, which makes it possible for TCP of classes I and II to compete for the sites of binding. TCP proteins participate in transduction of signals of hormonal origin and are redox-sensitive regulators of mitochondrial components of ETC encoded in the nucleus; they play an important role in mitochondrial biogenesis [67].
Not only АОХ expression is regulated by hormones but signal pathways of АОХ activation intercross hormonal signaling. A. thaliana mutants with an impaired polar transport of auxin (rao3/big, rao4/pin-formed1, and rao5/multidrug-resistance1/abcb19) showed more intense expression of АОХ1а [67]. Induction of expression of firefly luciferase reporter gene controlled by the AOX1a promoter was suppressed by auxin analogs and activated by prohibition of hormone transport. In addition, antimycin inhibited transduction of auxin signals and induced expression of АОХ1а, and auxin treatment partially eliminated this effect. This points to an antagonistic relationship between MRR signaling and kinetics of cell response to auxin. It was shown that cell response to auxin may be positively regulated by mitochondrial function via TCP. It was assumed that transduction of mitochondrial stress signal and auxin signaling are regulated reciprocally counterbalancing growth and stress response of the organism [67].
Light is a source of energy for all plants. The cells perceive light signals by means of photoreceptors (phytochromes, phototropins, cryptochromes, etc.) [71]. Anterograde signal cascades causing changes in the expression of light-regulated genes start with activation of photoreceptor and its migration to the nucleus (in the case of phytochromes). In the nucleus, the activated photoreceptor interacts with the Е3-ubiquitin-ligase complex СОР1/SPA1 that releases TF HY5, which initiates transcription of target genes. Phytochromes can directly interact with phytochrome-inactivating factors (for instance, with PIF3) containing a domain of DNA binding characteristic of transcription factors. Transduction of a light signal perceived by photoreceptors may involve secondary messengers: Ca2+, cAMP, cGMP, G-proteins, and other components. Phytochrome А is considered to play a key role in de-etiolation and photomorphogenesis, especially at insufficient illumination, while phytochrome B is considered to play a key role in plant response to elevated illuminance, although both phytochromes are important for the process of photomorphogenesis [71].
At present, light regulation of АОХ expression is being actively investigated [7, 32, 36, 44, 72]. AOX is considered to participate in oxidation of excess NADPH of a chloroplastic origin promoting the unloading of chlETC [73, 74]. It was shown that light regulation of АОХ1а expression in A. thaliana may involve phytochromes А and B, cryptochromes 1 and 2, and phototropins 1 and 2 [31, 75]. A signal from photoreceptors to the stage of АОХ transcription is transduced by means of known mechanisms of phototransduction of light-regulated (and first of all, photosynthetic) genes [76]. Light-dependent nature of АОХ1а induction was revealed in the leaves of A. thaliana [36, 51, 75] and Triticum aestivum [32], AOX2a in cotyledons of Glycine max [77], and АОХ1с in the leaves of Oryza sativa [44].
Analysis of a 1000-bp-long AtAOX1a promoter upstream of TSS revealed 27 light-dependent CAREs; out of them, nine were strictly light-induced [31]. The promoter of gene АОХ1а from A. thaliana contained a common G-box motif (CACGTG) [78] and CARE I1 (ACGTG, –93 bp from TSS) overlapping G-box [26]. It is known that G-box is a palindrome conserved CARE found in promoters of the majority of light-regulated genes; it is a binding site of GBFs transcription factors (G-box-Binding Factors) of family bZIP [79]. Light-induced transport of GBFs (in particular, of GBF-1) from cytoplasm to the nucleus is an important stage of signal transduction between photoreception and light-dependent gene expression. G-box was also found in the promoter of АОХ1а in Triticum urartu (GenBank: KD149474.1, –221 and –340 bp from TSS), a diploid progenitor of allohexaploid Triticum aestivum [32]. Analysis of available data suggests that there exists a mechanism connecting АОХ1а (at least in A. thaliana and Triticum aestivum) with photosynthetic function. Thus, investigation of the AtAOX1a promoter made it possible to detect light-induced CAREs controlled by yet unknown TFs and element I1 probably responsible for the connection between АОХ and photosynthesis. The role of different CAREs in the АОХ1а promoter and TFs recognizing them in light-dependent regulation of AOX1a expression is to be investigated.
On the basis of averaged values of gene expression observed in the course of greening relative to gene expression in etiolated wheat leaves, we proposed a pattern describing the induction of АОХ and the genes of other respiratory components of energy-dissipating ETC systems in the course of wheat leaf greening [7]. External and internal alternative NADPH-dehydrogenases participate in electron transport to ubiquinone bypassing complex I. The systems dissipating the proton gradient generated in the course of ETC reactions involve uncoupling proteins (UCP) similar to animal UCP. Via phytochrome-mediated pathway, light induces expression of АОХ1а and subsequent participation of AOX in respiration. In the period of fast development of the thylakoid system, this promoted de novo syntheses and prevented oxidative stress [32]. After 6-h-long greening, the weakening of AP modified the redox state of ETC and produced a redox signal transduced from mitochondria to the nucleus along the retrograde pathway, which initiated expression of the genes of other respiratory nonphosphorylating pathways and antioxidant enzymes participating in the maintenance of redox balance in the cell. In 24 h, the signals from mature chloroplasts [80] arrived in the nucleus and regulated expression of respective genes [32]. They were probably biogenic signals whose main member was protein GUN1 (Genome UNcopled1) located in the chloroplasts and responsible for homeostasis of plastid proteins. GUN1 is assumed to generate and transduce an unknown signal activating TF ABI4 that subsequently down-regulates expression of nuclear photosynthetic genes [81, 82]. GUN1 is believed to integrate numerous plastid signals related to the state of tetrapyrroles, chlETC, and redox state of plastids as a whole. Participation of GUN1/ABI4-signal pathway in the regulation of expression of АОХ1а and other respiratory genes in the course of formation of the photosynthetic apparatus is absolutely unexplored.
At the same time, it is not ruled out that MRR-dependent pathway of AOX1a induction plays a role in the process of greening, specifically via changes in the content of organic acids involved in the TCA cycle [7]. We observed an intensification of mitochondrial respiration and a rise in potential activity of AP in the presence of malate during the first 6 h of de-etiolation, which could point not only to a more active operation of ETC complex I at that time but also to activation of malate dehydrogenase of the TCA cycle [7]. It is known that the TCA cycle continues operating in the light with some modifications [83]. Changes in the content of organic acids participating in the TCA cycle (citrate, malate, and 2-oxoglutarate) were shown to affect the expression of AOX genes [68]. Specifically, when the TCA cycle is inhibited, AtAOXla signaling (the same as ROS-dependent pathway) involves the MRR region of the promoter. WRKY transcription factors were reported to play a role in AtAOXla response to MFA, which inhibits aconitase of the TCA cycle and leads to the accumulation of citrate [28]. It should be noted that participation of the ROS-dependent MRR pathway of signal transduction controlling АОХ1а in the course of wheat leaf greening is less evident. It is known that etiolated cells are protected from excessive ROS formation by the operation of light-dependent NADPH-protochlorophyllide oxidoreductase (LPOR); arising chlorophyllide-LPOR-NADH ternary complexes are powerful quenchers of energy [84]. In our investigation, the formation of ROS in wheat leaves even decreased in early stages of chloroplast biogenesis [32].
Our data suggest that the alternative pathway of respiration via AOX operates as a modulator of mitochondrial function during de-etiolation, and AOX plays a role in coordination of involvement of other respiratory nonphosphorylating pathways (NPP) in order to improve stress response and strengthen the stress signal. The idea that AOX participates in the coordination of other respiratory pathways and cellular defense systems was shared by other authors [1, 60]. Investigations conducted with АОХ1а-mutants of A. thaliana have shown that deficient AOX (either in gene expression or in the quantity of protein) in contrast to the components of other NPP caused modification of expression profiles of numerous genes and the whole transcriptome [1, 11, 36, 85]. There are also some reports that corroborate АОХ1а-dependent expression of genes from other NPP [51, 56] but show that АОХ1а does not depend on these genes [86]. A. thaliana plants with concurrent overexpression of АОХ1а and NDB2 easily survived drought and high illuminance, the same as the plants with overexpression of only one gene (АОХ1а) in contrast to plants overexpressing NDB2 [87]. Promoters of the majority of NPP genes of A. thaliana have identical stress-sensitive CAREs with АОХ1а [26, 35, 36]. However, the existence and quantity of common motifs in promoters does not always correlate with gene expression, which points to only the potential ability of the genes to interact at the transcriptional level [35, 36, 51].
Chloroplast-dependent signal pathways of АОХ expression. Chloroplasts and mitochondria are closely related not only by metabolic reactions but also by interactions at the level of retrograde and anterograde signals [1, 13, 30]. This relationship makes it possible to avoid oxidative stress in the chloroplasts and in the whole photosynthesizing cell. Together with other components of ETC, AOX is considered to participate in oxidation of the excess reducing agents transported by means of different shuttle mechanisms from chloroplasts to mitochondria [73, 74]. ROS, redox status of ETC components (mainly, plastoquinones, PQ), and redox-active molecules of plastidal stroma (thioredoxin and glutathione), as well as intermediates of tetrapyrrole biosynthesis, act as signal molecules of plastid retrograde pathways controlling expression of nuclear target genes, including components of mitochondrial respiration [81, 82, 88].
Expression of АОХ is closely related to chloroplast status. In greening under intense light (500 µmol/(m2 s1)), leaves of A. thaliana with a knockout at AOX1a (aox1a) and in plants of wild ecotype treated with AOX inhibitor salicylhydroxamic acid, a decrease in NADP+/NADPH ratio was accompanied by a reduced efficiency of import of plastid protein [89]. On the contrary, overexpression of AOX1a induced accumulation of chlorophyll and intensified the import of plastid protein. Under moderate illumination (350 µmol/(m2 s)), the pool of ubiquinone (UQ) and PQ in aox1a mutants was already more reduced [90], and maximum quantum yield of photosystem II (Fv/Fm) had lower values than in wild type plants [91]. In Nicotiana tabacum L. mutants with different levels of АОХ1а expression, plant acclimation to intense light and drought aimed to avoid overreduction of the PQ pool by means of lowering the quantity of protein LHCB2 of light-harvesting complex II and elevating the content of AOX [92]. The authors assumed that a signal from chlETC regulated the quantity of both proteins promoting the maintenance of energy balance in chloroplasts under stress. The obtained data confirm that the redox signal quickly inducing АОХ expression arrives directly from chloroplasts and the level of PQ pool reduction is a redox sensor [1, 88].
Recent results suggest that mitochondrial retrograde signal pathways are related to the network of cell signals’ transduction and can integrate information from several sources, including chloroplast, and AOX actively participates in the maintenance of cell signaling and interaction between chloroplasts and mitochondria. Key TFs involved in regulation of AtАОХ1а expression (ABI4, ANAC013, ANAC017, WRKY40, and WRKY63) are components of plastid retrograde signaling [12, 34, 82]. The overexpression of ANAC017 improved the expression of АОХ but reduced the expression of the genes encoding chloroplast proteins, including LHCB [34]. In the dark, mitochondrial dysfunctions in the leaves of Nicotiana tabacum L. caused by different inhibitors improved the expression of MDS-genes, including АОХ1 genes [93]. However, expression of АОХ1 genes decreased in light under the effect of antimycin in contrast to myxothiazol, which did not reduce elevated gene expression. Antimycin (same as myxothiazol) is an inhibitor of ETC complex III but also inhibits ferredoxin-quinone reductase participating in electron transfer from the acceptor side of PS I to PQ in cyclic electron flow (CEF) around PS I of chlETC. It is believed that acceptor-side limitation of PS I is a source of chloroplast retrograde signals. At the same time, myxothiazol elevated NPQ and antimycin reduced this coefficient [93]. Thus, a decrease in АОХ1 expression in the presence of antimycin in light resulted from the interaction between the signals arriving from both organelles. It is interesting that inhibition of CEF by antimycin did not affect expression of other MDS-genes in the light in plants with overexpression of АОХ1а. In other words, the redox state of chloroplast actually affects expression of АОХ and other MDS-genes, but mitochondrial dysfunction accompanied by a disturbance of electron flow to oxygen is a primary condition for induction of gene expression. On the whole, the integration of signals is one of the instruments of coordination of metabolic interactions between organelles, which determine energy status of the cell. Most probably, a key signal of this integration is ROS generated by both organelles [1, 13].
It is assumed that overreduction of the UQ pool in a photosynthesizing cell, especially at intense illuminance, may also result from an increase in photorespiratory NADH and lead to a more intense formation of ROS and, therefore, to opening of PTP with subsequent induction of АОХ [66, 88]. Photosynthetic activity causes changes in the content of the main carbon metabolites (sugars, amino acids, and intermediates of TCA cycle) and reducing potential of pyridine nucleotides and protein thiol groups. This may presumably affect gene expression and the activity of mitochondrial proteins, including AOX [88]. It was shown that diurnal changes in the level of sugars (a decrease by the end of night and a rise by the midday), accompanied by changes in the content of different hormones (auxin and cytokinin by the end of night, ABA by the midday), control expression of numerous genes, including ETC carriers [76]. This suggests a hormone-dependent nature of transduction of signals regulating diurnal gene expression. Taking into consideration the fact that gene AtAOX1a promoter contains ABRE motifs, it is possible that this mechanism is also involved in light regulation of АОХ expression [26]. It was revealed that the activity of cytochrome and alternative pathways observed in vivo in double cytc (cytc 1b2a and cyt c1b2b) mutants of A. thaliana depended on the duration of the photoperiod that influenced operation of the TCA cycle, composition of its intermediates and amino acids and other metabolites [94]. Changes in metabolism gave signals about stress by triggering reactions modifying primary metabolism for adaptation to a different photoperiod. In particular, under a long day, cytc accumulated more gamma-aminobutyric acid (GABA) and showed a high activity of AOX. GABA can act as an internal stress signal in plants modifying the activity of ion transporters. In addition to a mediator effect, GABA directly participates in metabolism and turns into succinic acid (succinate) with subsequent entry into the TCA cycle. The so-called GABA shunt ensures an alternative supply of carbon to the TCA cycle in the form of succinate bypassing succinyl-CoA synthetase and diminishing the TCA cycle turnover, and involvement of AOX makes it possible to maintain redox status of ETC upon a dysfunction of the main cytochrome pathway. It is considered that cytochrome с as a key component of CP plays an important role in plant growth and development. Mutants cytc had a low level of gibberellins and were notable for an elevated content of DELLA proteins that repress gibberellin signaling. Therefore, a metabolic connection between GABA shunt and AOX activation is probably integrated into the network of hormonal signaling [94].
AOX PROTEIN AND POSTTRANSLATIONAL REGULATION OF ITS ACTIVITY
Gene expression shows in transcription of information from DNA to mRNA with subsequent translation into a sequence of the protein molecule. Alternative oxidase (EC 1.10.3.11), ubiquinol:oxygen oxidoreductase, is a homodimeric protein (mass of a single polypeptide is approximately 32 kD). The protein is anchored in one layer of inner mitochondrial membrane on the matrix side by means of its large hydrophobic fragment formed by helixes [95]. AOX contains a hydroxo-bridged di-iron binuclear center of mixed valency (Fe(II)/Fe(III)). Each iron center resides within a four-helix bundle; it is conserved for all isoforms of AOX and acts as an active site for reduction of oxygen to water. Domains of N-termini of AOX monomers are linked with a covalent disulphide bond at the expense of bridges formed between cysteine residues crucial for dimer structure [95].
AOX is produced in cyto-ribosomes as a protein precursor with an appropriate signal sequence at the N-terminus (presequence) [96]. Protein precursor of AOX is imported into mitochondria via specific translocation pores with the assistance of chaperons. After entry of the protein precursor into mitochondrion, the presequence is removed by mitochondrial processing peptidase [96].
Reduction of the disulphide bond to a sulfhydryl one results in activation of AOX. AOX activity in vitro (in isolated mitochondria) depends on redox state of the enzyme, substrate availability (extent of UQ reducibility), key metabolites (α-keto acids), and the quantity of synthesized protein. Mechanisms of AOX activation in vitro are investigated in detail [3–5]. The thioredoxin (Trx) system is responsible for reduction of AOX protein; in turn, it depends on redox state of the NADPH pool. Trx protein that has a disulfide bond uses electrons and protons from NADPH reducing AOX for its reduction. At the same time, redox state of the NADPH pool depends on the operation of NADP+-dependent isocitrate dehydrogenase [5].
The activity of reduced form of AOX remains low and increases when UQ reducibility is high and/or organic acids are available. The main metabolites activating reduced dimer of AOX are α-keto acids (pyruvate and glyoxylate). The interaction of cysteine residues with α-keto acid results in the formation of thiohemiacetal, a completely activated form of AOX [5].
Depending on the genes encoding AOX, there are different isoforms of AOX. The majority of isoforms have two cysteine residues but there are exceptions. Isoforms АОХ1А and АОХ1В of A. thaliana and AOX isoforms of some other species, for instance, of tomato and lotus, have the third cysteine residue located near the catalytic center containing an iron atom [5]. Analysis of posttranslational activation of AOX revealed specificity of protein isoforms in respect to metabolites activating AOX. АОХ1А was more sensitive to α-keto acids than АОХ1С and АОХ1D [97]. TCA cycle acids specifically activate AO isoforms. АОХ1А is stimulated by OA or 2-oxoglutarate (2-OG), while АОХ1D only by 2-ОG, and АОХ1С is insensitive to intermediates of the TCA cycle. At the same time, not all the isoforms of AOX contain cysteine residue at the first position. АОХ1В isoforms of tomato, lotus, and rice and АОХ3 of NCS-mutants of maize contain serine at this position and are insensitive to α-keto acids, but they are activated by succinate via the formation of an ester bond [59, 97]. Isoforms of АОХ2 are less investigated in respect to their activation by metabolites; however, available data show that the main activator of АОХ2 is pyruvate [5].
The level of expression of AOX genes does not always correspond to the quantity of AOX protein and the latter to capacity or activity of the enzyme in vivo (Supplementary Table S1, [6]). Moreover, AOX activity in vivo is often lower than its capacity (or maximum activity), which is related to limitation of substrate availability, allosteric activators (α-keto acids), and other mechanisms of posttranslational activation of the enzyme [6]. AP activity in vivo (valt) may be examined by the method of isotope (18О/16О) discrimination [6]. Capacity or maximum activity of AOX (Valt) is determined by the method of specific inhibitors, which is currently widely used [3, 6]. AOX inhibitors are hydroxamic acids and n-propyl gallate. Valt value is determined by the sensitivity of respiration to inhibitor of AOX and inhibitor of cytochrome oxidase cyanide (CN–) less residual respiration (in the presence of both inhibitors). Optimal concentrations of inhibitors are determined by titration of the sample with increasing concentrations of inhibitors up to the saturation of О2 uptake rate [3]. AP respiration is sometimes designated as cyanide-resistant respiration (CRR): respiration in the presence of CP inhibitor alone. CRR does not reflect Valt value correctly.
Investigations often show a higher AOX capacity as compared with AOX activity in vivo and lack of changes in the level of AOX protein upon exposure to stress factors, which is apparently associated with the maintenance of capacity for fast activation of the enzyme bypassing energy-consuming processes of de novo protein synthesis [6, 76]. This makes respiratory metabolism flexible and adaptable and allows the cell to rapidly respond to internal and environmental changes.
At the same time, the data corroborating the existence of mechanisms of posttranslational regulation of AOX in vivo are not numerous. In plants genetically modified by АОХ, it was shown that the level of AOX protein is not the main factor of regulation of enzyme activity in vivo but can affect in vivo AOX activity under stress. In particular, under normal controlled growing conditions, the leaves of tobacco plants with an elevated content of AOX protein (line S24) had a greater value of CRR in contrast to antisense line AS8 with a reduced content of AOX protein [98]. At the same time, all the lines, including wild type, had similar AP activity in vivo. However, under different types of stress (drought and photoinhibition), the leaves of line S24 (in contrast to wild type) showed an elevated activity of AP in vivo. In another paper, a brief exposure to high illuminance (800 µmol/(m2 s)) resulted in elevation of AOX activity in vivo in the leaves of line ХХ-2 with a higher level of AOX protein; in antisense by АОХ1а plants of A. thaliana (line AS-12), it did not change [99]. It is interesting that the inability to increase the rate of AP respiration in the leaves of line AS-12 under these conditions was compensated for by activation of CP respiration. This suggests that changes in metabolic processes and/or biochemical mechanisms of regulation of AOX protein activity are crucial for electron partitioning to AP.
The role of AOX in maintenance of the ubiquinone pool has been discussed for a long time, but convincing experimental arguments that the ubiquinone pool (or rather the ratio between reduced and total ubiquinone, Qr/Qt) is the factor regulating AOX activity in vivo are still lacking [4]. Available information only indirectly confirms this idea. Specifically, at a short photoinhibition, the leaves of antisense by АОХ1а line of A. thaliana showed an elevated Qr/Qt ratio when AOX in vivo was not involved and CP was saturated concurrently [99]. It is also known that reducibility of the ubiquinone pool is partially regulated by NADPH-dehydrogenase; at the same time, some genes encoding NADPH-dehydrogenases are coexpressed with AOX genes [35, 36, 51, 76, 78]. These scanty data suggest that reducibility of the ubiquinone pool, depending on NADH concentration in the cytosol and mitochondria, affects the activity of AOX in vivo [6]. An opinion was expressed that activation of AOX in vivo requires only little changes in Qr/Qt value [4]. Existence of the mechanisms of posttranslational regulation of AOX in vivo by α-keto acids and the Trx system controlling redox state of the enzyme has not been corroborated experimentally and requires further investigation.
CONCLUSIONS
Mitochondrial alternative oxidase is a key enzyme that directly participates in regulation of energy and redox balance of mitochondria and the whole cell. Available literature data show various mechanisms and intricate combinations of signal pathways regulating expression of AOX genes. AOX can directly affect stress signaling via reduction of ROS generation and indirectly via modification of cell energy status and triggering certain signal cascades. It is interesting that numerous mechanisms of transcriptional and posttranscriptional control operate in the cell; they ensure a suppression of AOX genes’ expression in normal conditions and a rapid induction of their expression under stress (by the example of AtAOX1a). AOX is encoded by several genes, which makes it possible to produce different forms of protein with different mechanisms of posttranslational activation. This improves protection of the function performed by the protein in a plant cell. At present, mitochondrial retrograde regulation of АОХ expression is studied in detail. Indistinct differences between mitochondrial and plastidal retrograde signal pathways emphasize a close relationship between organelles but complicate investigation of the role of AOX signaling in control over energy metabolism of the photosynthesizing cell. It became evident that signal pathways of regulation of АОХ expression overlap the hormonal system of signaling. There appeared innumerous reports indicating that AOX is not only a target but also a regulator of stress signal; it coordinates expression of other ETC genes and generates and/or amplifies stress signal. All these issues require further investigation. Before long, accumulation of knowledge about AOX signaling will ensure their practical application in an effort to achieve ecological, clinical, and agricultural purposes.
REFERENCES
Vanlerberghe, G.C., Dahal, K., Alber, N.A., and Chadee, A., Photosynthesis, respiration and growth: a carbon and energy balancing act for alternative oxidase, Mitochondrion, 2020, vol. 52, p. 197.
Meeuse, B.J.D., Thermogenic respiration in aroids, Plant Physiol., 1975, vol. 26, p. 117.
Shugaev, A.G., Alternative cyanide-resistant oxidase in plant mitochondria: structure, regulation of activity, and presumable physiological role, Russ. J. Plant Physiol., 1999, vol. 46, p. 262.
Millenaar, F.F. and Lambers, H., The Alternative oxidase: in vivo regulation and function, Plant Biol., 2003, vol. 5, p. 2.
Selinski, J., Scheibe, R., Day, D.A., and Whelan, J., Alternative oxidase is positive for plant performance, Trends Plant Sci., 2018, vol. 23, p. 588.
Del-Saz, N.F., N., Ribas-Carbo, M., McDonald, A.E., Lambers, H., Fernie, A.R., and Florez-Sarasa, I., An in vivo perspective of the role(s) of the alternative oxidase pathway, Trends Plant Sci., 2018, vol. 23, p. 206.
Garmash, E.V., Role of mitochondrial alternative oxidase in the regulation of cellular homeostasis during development of photosynthetic function in greening leaves, Plant Biol., 2021, vol. 23, p. 221.
Arnholdt-Schmitt, B., Costa, J.H., and Fernandes de Melo, D., AOX—a functional marker for efficient cell reprogramming under stress? Trends Plant Sci., 2006, vol. 11, p. 281.
González-Meler, M.A., Blanc-Betes, E., Flower, C.E., and Ward, J.K., Plastic and adaptive responses of plant respiration to changes in atmospheric CO2 concentration, Physiol. Plant., 2009, vol. 137, p. 473.
Saari, S., Garcia, G.S., Bremer, K., Chioda, M.M., Andjelkovic, A., Debes, P.V., Nikinmaa, M., Szibor, M., Dufour, E., Rustin, P., Oliveira, M.T., and Jacobs, H.T., Alternative respiratory chain enzymes: therapeutic potential and possible pitfalls, Biochim. Biophys. Acta, Mol. Basis Dis., 2018, vol. 1865, p. 854.
Van Aken, O., Giraud, E., Clifton, R., and Whelan, J., Alternative oxidase: a target and regulator of stress responses, Physiol. Plant., 2009, vol. 137, p. 354.
Ng, S., De Clercq, I., van Aken, O., Law, S.R., Ivanova, A., Willems, P., Giraud, E., van Breusegem, F., and Whelan, J., Anterograde and retrograde regulation of nuclear genes encoding mitochondrial proteins during growth, development, and stress, Mol. Plant., 2014, vol. 7, p. 1075.
Van Aken, O., Mitochondrial redox systems as central hubs in plant metabolism and signaling, Plant Physiol., 2021, vol. 186, p. 36.
Polidoros, A.N., Mylona, P.V., and Arnholdt-Schmitt, B., Aox gene structure, transcript variation and expression in plants, Physiol. Plant., 2009, vol. 137, p. 342.
Costa, J.H., Pereira dos Santos, C., de Sousa e Lima, B., Moreira Netto, A. N., da Cruz Saraiva, K.D., and Arnholdt-Schmitt, B., In silico identification of alternative oxidase 2 (AOX2) in monocots: a new evolutionary scenario, J. Plant Physiol., 2017, vol. 210, p. 58.
Takumi, S., Tomioka, M., Eto, K., Naydenov, N., and Nakamura, C., Characterization of two non-homoeologous nuclear genes encoding mitochondrial alternative oxidase in common wheat, Genes Genet. Syst., 2002, vol. 77, p. 81.
Brew-Appiah, R.A.T., York, Z.B., Krishnan, V., Roalson, E., and Sanguinet, K.A., Genome-wide identification and analysis of the ALTERNATIVE OXIDASE gene family in diploid and hexaploid wheat, PLoS One, 2018, vol. 13, p. e0201439. https://doi.org/10.1371/journal.pone.0201439
Campos, M.D., Cardoso, H.G., Linke, B., Costa, J.H., Fernandes de Melo, D., Justo, L., Frederico, A.M., and Arnholdt-Schmitt, B., Differential expression and co-regulation of carrot AOX genes (Daucus carota), Physiol. Plant., 2009, vol. 137, p. 578.
Macedo, E.S., Cardoso, H.G., Hernandez, A., Peixe, A.A., Polidoros, A., Ferreira, A., Cordeiro, A., and Arnholdt-Schmitt, B., Physiologic responses and gene diversity indicate olive alternative oxidase as a potential source for markers involved in efficient adventitious root induction, Physiol. Plant., 2009, vol. 137, p. 532.
Cardoso, H.G., Campos, M.D., Costa, A.R., Campos, M.C., Nothnagel, T., and Arnholdt-Schmitt, B., Carrot alternative oxidase gene AOX2a demonstrates allelic and genotypic polymorphisms in intron 3, Physiol. Plant., 2009, vol. 137, p. 592.
Ding, C., Chen, C., Su, N., Lyu, W., Yang, J., Hu, Z., and Zhang, M., Identification and characterization of a natural SNP variant in ALTERNATIVE OXIDASE gene associated with cold stress tolerance in watermelon, Plant Sci., 2021, vol. 304, p. 110735 https://doi.org/10.1016/j.plantsci.2020.110735
Arnholdt-Schmitt, B., Efficient cell reprogramming as a target for functional-marker strategies? Towards new perspectives in applied plant-nutrition research, J. Plant Nutr. Soil Sci., 2005, vol. 168, p. 617.
Abe F., Saito K., Miura K., and Toriyama K., A single nucleotide polymorphism in the alternative oxidase gene among rice varieties differing in low temperature tolerance, FEBS Lett., 2002. Vol. 527, p. 181.
Polidoros, A.N., Mylona, P.V., Pasentsis, K., Scandalios, J.G., and Tsaftaris, A.S., The maize alternative oxidase 1a (Aox1a) gene is regulated by signals related to oxidative stress, Redox Rep., 2005, vol. 10, p. 71.
Ho, L.H.M., Giraud, E., Lister, R., Thirkettle-Watts, D., Low, J., Clifton, R., Howell, K.A., Carrie, C., Donald, T., and Whelan, J., Characterization of the regulatory and expression context of an alternative oxidase gene provides insights into cyanide-insensitive respiration during growth and development, Plant Physiol., 2007, vol. 143, p. 1519.
Ho, L.H.M., Giraud, E., Uggalla, V., Lister, R., Clifton, R., Glen, A., Thirkettle-Watts, D., van Aken, O., and Whelan, J., Identification of regulatory pathways controlling gene expression of stress-responsive mitochondrial proteins in Arabidopsis, Plant Physiol., 2008, vol. 147, p. 1858.
Thirkettle-Watts, D., McCabe, T.C., Clifton, R., Moore, C., Finnegan, P.M., Day, D.A., and Whelan, J., Analysis of the alternative oxidase promoters from soybean, Plant Physiol., 2003, vol. 133, p. 1158.
Dojcinovic, D., Krosting, J., Harris, A.J., Wagner, D.J., and Rhoads, D.M., Identification of a region of the Arabidopsis AtAOX1a promoter necessary for mitochondrial retrograde regulation of expression, Plant Mol. Biol., 2005, vol. 58, p. 159.
De Clercq, I., Vermeirssen, V., van Aken, O., Vandepoele, K., Murcha, M.W., Law, S.R., Inzé, A., Ng, S., Ivanova, A., Rombaut, D., van de Cotte, D., Jaspers, P., van de Peer, Y., Kangasjärvi, J., Whelan, J., and van Breusegem, F., The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis, Plant Cell, 2013, vol. 25, p. 3472.
Giraud, E., van Aken, O., Ho, L.H.M., and Whelan, J., The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a, Plant Physiol., 2009, vol. 150, p. 1286.
Zhang, D.-W., Xu, F., Zhang, Z.-W., Chen, Y.-E., Du, J.-B., Jia, S-D., Yuan, S., and Lin, H.-H., Effects of light on cyanide-resistant respiration and alternative oxidase function in Arabidopsis seedlings, Plant Cell Environ., 2010, vol. 33, p. 2121.
Garmash, E.V., Velegzhaninov, I.O., Grabelhych, O.I., Borovik, O.A., Silina, E.V., Voinikov, V.K., and Golovko, T.K., Expression profiles of genes for mitochondrial respiratory energy-dissipating systems and antioxidant enzymes in wheat leaves during de-etiolation, J. Plant Physiol., 2017, vol. 215, p. 110.
Van Aken, O., Zhang, B., Law, S., Narsai, R., and Whelan, J., AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins, Plant Physiol., 2013, vol. 162, p. 254.
Ng, S., Ivanova, A., Duncan, O., Law, S.R., Van Aken, O., De Clercq, I., Wang, Y., Carrie, C., Xu, L., Kmiec, B., Walker, H., Van Breusegem, F., Whelan, J., and Giraud, E., A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis, The Plant Cell, 2013, vol. 25, p. 3450.
Garmash, E.V., Velegzhaninov, I.O., Ermolina, K.V., Rybak, A.V., and Malyshev, R.V., Altered levels of AOX1a expression result in changes in metabolic pathways in Arabidopsis thaliana plants acclimated to low dose rates of ultraviolet B radiation, Plant Sci., 2020, vol. 291, p. 110332. https://doi.org/10.1016/j.plantsci.2019.110332
Garmash, E.V., Belykh, E.S., and Velegzhaninov, I.O., The gene expression profiles of mitochondrial respiratory components in Arabidopsis plants with differing amounts of ALTERNATIVE OXIDASE1a under high intensity light, Plant Signaling Behav., 2021, vol. 16, p. 3. https://doi.org/10.1080/15592324.2020.1864962
Nagel, D.H., Doherty, C.J., Pruneda-Paz, J.L., Schmitz, R.J., Ecker, J.R., and Kay, S.A., Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis, Proc. Natl. Acad. Sci. U.S.A., 2015, vol. 112, p. E4802-10. https://doi.org/10.1073/pnas.1513609112
Safrany, J., Haasz, V., Mate, Z., Ciolfi, A., Feher, B., Oravecz, A., Stec, A., Dallmann, G., Morelli, G., Ulm, R., and Nagy, F., Identification of a novel cis-regulatory element for UV-B-induced transcription in Arabidopsis, Plant J., 2008, vol. 54, p. 402.
Rhoads, D.M. and McIntosh, L., Salicylic acid regulation of respiration in higher plants: alternative oxidase expression, Plant Cell, 1992, vol. 4, p. 1131.
Ng, S., Giraud, E., Duncan, O., Law, S.R., Wang, Y., Xu, L., Narsai, R., Carrie, C., Walker, H., Day, D.A., Blanco, N.E., Strand, A., Whelan, J., and Ivanova, A., Cyclin-dependent kinase E1 (CDKE1) provides a cellular switch in plants between growth and stress responses, J. Biol. Chem., 2013, vol. 288, p. 3449.
Rhoads, D.M. and McIntosh, L., Isolation and characterization of a cDNA clone encoding an altemative oxidase protein of Sauromatum guttatum (Schott), Proc. Natl. Acad. Sci. U.S.A., 1991, vol. 88, p. 2122.
Clifton, R., Millar, A.H., and Whelan, J., Alternative oxidases in Arabidopsis: a comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses, Biochim. Biophys. Acta, Bioenerg., 2006, vol. 1757, p. 730.
Costa, J.H., Hasenfratz-Sauder, M.P., Pham-Thi, A.T., Lima, M.D.S., Dizengremel, P., Jolivet, Y., and de Melo, D.F., Identification in Vigna unguiculata (L.) Walp. of two cDNAs encoding mitochondrial alternative oxidase orthologous to soybean alternative oxidase genes 2a and 2b, Plant Sci., 2004, vol. 167, p. 233.
Feng, H.Q., Li, H.Y., Zhou, G.M., Liang, H.G., Duan, J.G., Zhi, D.J., Li, X., and Ma, J., Influence of irradiation on cyanide-resistant respiration and AOX1 multi-gene family expression during greening of etiolated rice seedlings, Photosynthetica, 2007, vol. 45, p. 272.
Garmash, E.V., Grabelhych, O.I., Velegzhaninov, I.O., Borovik, O.A., Dalke, I.V., Voinikov, V.K., and Golovko, T.K., Light regulation of AOX pathway during greening of etiolated wheat seedlings, J. Plant Physiol., 2015, vol. 174, p. 75.
Mizuno, N., Sugie, A., Kobayashi, F., and Takumi, S., Mitochondrial alternative pathway is associated with development of freezing tolerance in common wheat, J. Plant Physiol., 2008, vol. 165, p. 462.
Grabelnych, O.I., Borovik, O.A., Tauson, E.L., Pobezhimova, T.P., Katyshev, A.I., Pavlovskaya, N.S., Koroleva, N.A., Lyubushkina, I.V., Bashmakov, V.Yu., Popov, V.N., Borovskii, G.B., and Voinikov, V.K., Mitochondrial energy-dissipating systems (alternative oxidase, uncoupling proteins, and external NADH dehydrogenase) are involved in development of frost-resistance of winter wheat seedlings, Biochemistry (Moscow), 2014, vol. 79, p. 506.
Smith, C.A., Melino, V.J., Sweetman, C., and Soole, K.L., Manipulation of alternative oxidase can influence salt tolerance in Arabidopsis thaliana, Physiol. Plant., 2009, vol. 137, p. 459.
Wang, J. and Vanlerberghe, G.C., A lack of mitochondrial alternative oxidase compromises capacity to recover from severe drought stress, Physiol. Plant., 2013, vol. 149, p. 461.
Gupta, K.J., Shah, J.K., Brotman, Y., Jahnke, K., Willmitzer, L., Kaiser, W.M., Bauwe, H., and Igamberdiev, A.U., Inhibition of aconitase by nitric oxide leads to induction of the alternative oxidase and to a shift of metabolism towards biosynthesis of amino acids, J. Exp. Bot., 2012, vol. 63, p. 1773.
Yoshida, K. and Noguchi, K., Differential gene expression profiles of the mitochondrial respiratory components in illuminated Arabidopsis leaves, Plant Cell Physiol., 2009, vol. 50, p. 1449.
Zhu, L.-J., Deng, X.-G., Zou, L.-J., Zhang, D.-W., and Lin, H.-H., Enhancement of stress tolerance in cucumber seedlings by proanthocyanidins, Biol. Plant, 2017, vol. 61, p. 323.
Panda, S.K., Sahoo, L., Katsuhara, M., and Matsumoto, H., Overexpression of alternative oxidase gene confers aluminum tolerance by altering the respiratory capacity and the response to oxidative stress in tobacco cells, Mol. Biotechnol., 2012, vol. 54, p. 551.
Keunen, E., Jozefczak, M., Remans, T., Vangronsveld, J., and Cuypers, A., Alternative respiration as a primary defense during cadmium-induced mitochondrial oxidative challenge in Arabidopsis thaliana, Environ. Exp. Bot., 2013, vol. 91, p. 63.
Pasqualini, S., Paolocci, F., Borgogni, A., Morettini, R., and Ederli, L., The overexpression of an alternative oxidase gene triggers ozone sensitivity in tobacco plants, Plant Cell Environ., 2007, vol. 30, p. 1545.
Watanabe, C.K., Hachiya, T., Takahara, K., Kawai-Yamada, M., Uchimiya, H., Uesono, Y., Terashima, I., and Noguchi, K., Effects of AOX1a deficiency on plant growth, gene expression of respiratory components and metabolic profile under low-nitrogen stress in Arabidopsis thaliana, Plant Cell Physiol., 2010, vol. 51, p. 810.
Cvetkovska, M. and Vanlerberghe, G.C., Alternative oxidase impacts the plant response to biotic stress by influencing the mitochondrial generation of reactive oxygen species, Plant Cell Environ., 2013, vol. 36, p. 721.
Matos, A.R., Mendesa, A.T., Scotti-Campos, P., and Arrabaca, J.D., Study of the effects of salicylic acid on soybean mitochondrial lipids and respiratory properties using the alternative oxidase as a stress-reporter protein, Physiol. Plant., 2009, vol. 137, p. 485.
Karpova, O.V., Kuzmin, E.V., Elthon, T.E., and Newton, K.J., Differential expression of alternative oxidase genes in maize mitochondrial mutants, Plant Cell, 2002, vol. 14, p. 3271.
Juszczuk, I.M., Szal, B., and Rychter, A.M., Oxidation-reduction and reactive oxygen species homeostasis in mutant plants with respiratory chain complex I dysfunction, Plant Cell Environ., 2012, vol. 35, p. 296.
Yurina, N.P. and Odintsova, M.S., Signal transduction pathways of plant mitochondria: retrograde regulation, Russ. J. Plant Physiol., 2010, vol. 57, p. 7.
Li, C.R., Liang, D.D., Xu, R.F., Li, H., Zhang, Y.P., Qin, R.Y., Li, L., Wei, P.C., and Yang, J.B., Overexpression of an alternative oxidase gene, OsAOX1a, improves cold tolerance in Oryza sativa L., Genet. Mol. Res., 2013, vol. 12, p. 5424.
Liu, J., Li, Z., Wang, Y., and Xing, D., Overexpression of ALTERNATIVE OXIDASE1a alleviates mitochondria-dependent programmed cell death induced by aluminum phytotoxicity in Arabidopsis, J. Exp. Bot., 2014, vol. 65, p. 4465.
Wagner, A.M., A role for active oxygen species as second messengers in the induction of alternative oxidase gene expression in Petunia hybrida cells, FEBS Lett., 1995, vol. 368, p. 339.
Maxwell, D.P., Nickels, R., and McIntosh, L., Evidence of mitochondrial involvement in the transduction of signals required for the induction of genes associated with pathogen attack and senescence, Plant J., 2002, vol. 29, p. 269.
Amirsadeghi, S., Robson, C.A., and Vanlerberghe, G.C., The role of the mitochondrion in plant responses to biotic stress, Physiol. Plant., 2007, vol. 129, p. 253.
Ivanova, A., Law, S.R., Narsai, R., Duncan, O., Lee, J.H., Zhang, B., van Aken, O., Radomiljac, J.D., van der Merwe, M., Yi, K., and Whelan, J.A., Functional antagonistic relationship between auxin and mitochondrial retrograde signaling regulates Alternative Oxidase1a expression in Arabidopsis, Plant Physiol., 2014, vol. 165, p. 1233.
Gray, G.R., Maxwell, D.P., Villarimo, A.R., and McIntosh, L., Mitochondria/nuclear signaling of alternative oxidase gene expression occurs through distinct pathways involving organic acids and reactive oxygen species, Plant Cell Rep., 2004, vol. 23, p. 497.
Robson, C.A. and Vanlerberghe, G.C., Transgenic plant cells lacking mitochondrial alternative oxidase have increased susceptibility to mitochondria-dependent and -independent pathways of programmed cell death, Plant Physiol., 2002, vol. 129, p. 1908.
Vanderauwera, S., Vandenbroucke, K., Inze, A., van de Cotte, B., Muhlenbock, P., De Rycke, R., Naouar, N., van Gaever, T., van Montagu, M.C., and van Breusegem, F., AtWRKY15 perturbation abolishes the mitochondrial stress response that steers osmotic stress tolerance in Arabidopsis, Proc. Natl. Acad. Sci. U.S.A., 2012, vol. 109, p. 20113.
Kreslavski, V.D., Carpentier, R., Klimov, V.V., and Allakhverdiev, S.I., Transduction mechanisms of photoreceptor signals in plant cells, J. Photochem. Photobiol., C, 2009, vol. 10, p. 63.
Ribas-Carbó, M., Aroca, R., Gonzàlez-Meler, M.A., Irigoyen, J.J., and Sanchez-Diaz, M., The electron partitioning between the cytochrome and alternative respiratory pathways during chilling recovery in two cultivars of maize differing in chilling sensitivity, Plant Physiol., 2000, vol. 122, p. 199.
Noguchi, K. and Yoshida, K., Interaction between photosynthesis and respiration in illuminated leaves, Mitochondrion, 2008, vol. 8, p. 887.
Dinakar, C., Raghavendra, A.S., and Padmasree, K., Importance of AOX pathway in optimizing photosynthesis under high light stress: role of pyruvate and malate in activating of AOX, Physiol. Plant, 2010, vol. 139, p. 13.
Tepperman, J.M., Hudson, M.E., Khanna, R., Zhu, T., Chang, S.H., Wang, X., and Quail, P.H., Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation, Plant J., 2004, vol. 38, p. 725.
Rasmusson, A.G. and Escobar, M., Light and diurnal regulation of plant respiratory gene expression, Physiol. Plant., 2007, vol. 129, p. 57.
Finnegan, P.M., Whelan, J., Millar, A.H., Zhang, Q., Smith, M.K., Wiskich, J.T., and Day, D.A., Differential expression of the multigene family encoding the soybean mitochondrial alternative oxidase, Plant Physiol., 1997, vol. 114, p. 455.
Clifton, R., Lister, R., Parker, K.L., Sappl, P.G., Elhafez, D., Millar, A.H., Day, D.A., and Whelan, J., Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana, Plant Mol. Biol., 2005, vol. 58, p. 193.
Menkens, A.E., Schindler, U., and Cashmore, A.R., The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins, Trends Biochem Sci., 1995, vol. 20, p. 506.
Garmash, E.V., Dymova, O.V., Malyshev, R.V., Plyusnina, S.N., and Golovko, T.K., Developmental changes in energy dissipation in etiolated wheat seedlings during the greening process, Photosynthetica, 2013, vol. 51, p. 497.
Kusnetsov, V.V., Chloroplasts: structure and expression of the plastid genome, Russ. J. Plant Physiol., 2018, vol. 65, p. 465.
Yurina, N.P. and Odintsova, M.S., Chloroplast retrograde signaling system, Russ. J. Plant Physiol., 2019, vol. 66, p. 509.
Tcherkez, G., Boex-Fontvieille, E., Mahe, A., and Hodges, M., Respiratory carbon fluxes in leaves, Curr. Opin. Plant Biol., 2012, vol. 15, p. 308.
Schoefs, B. and Franck, F., Protochlorophyllide reduction: mechanisms and evolution, Photochem. Photobiol., 2003, vol. 78, p. 543.
Wallström, S.V., Florez-Sarasa, I., Wagner, L., Ara-újo, W.L., Escobar, M.A., Geisler, D.A., Aidemark, M., Lager, I., Fernie, A.R., Ribas-Carbo, M., and Rasmusson, A.G., Suppression of NDA-type alternative mitochondrial NAD(P)H dehydrogenases in Arabidopsis thaliana modifies growth and metabolism, but not high light stimulation of mitochondrial electron transport, Plant Cell Physiol., 2014, vol. 55, p. 881.
Sweetman, C., Waterman, C.D., Rainbird, B.M., Smith, P.M.C., Jenkins, C.D., Day, D.A., and Soole, K.L., AtNDB2 is the main external NADH dehydrogenase in mitochondria and is important for tolerance to environmental stress, Plant Physiol., 2019, vol. 181, p. 774.
Giraud, E., Ho, L.H.M., Clifton, R., Carroll, A., Estavillo, G., Tan, Y.F., Howell, K.A., Ivanova, A., Pogson, B.J., Millar, A.H., and Whelan, J., The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress, Plant Physiol., 2008, vol. 147, p. 595.
Van Lis, R. and Atteia, A., Control of mitochondrial function via photosynthetic redox signals, Photosynth. Res., 2004, vol. 79, p. 133.
Zhang, D.-W., Yuan, S., Xu, F., Zhu, F., Yuan, M., Ye, H.-X., Guo, H.-Q., Lv, X., Yin, Y., and Lin, H.-H., Light intensity affects chlorophyll synthesis during greening process by metabolite signal from mitochondrial alternative oxidase in Arabidopsis, Plant Cell Environ., 2016, vol. 39, p. 12.
Yoshida, K., Watanabe, C.K., Hachiya, T., Tholen, D., Shibata, M., Terashima, I., and Noguchi, K., Distinct responses of the mitochondrial respiratory chain to long- and short-term high-light environments in Arabidopsis thaliana, Plant Cell Environ., 2011, vol. 34, p. 618.
Watanabe, C.K.A., Yamori, W., Takahashi, S., Terashima, I., and Noguchi, K., Mitochondrial alternative pathway-associated photoprotection of photosystem II is related to the photorespiratory pathway, Plant Cell Physiol., 2016, vol. 57, p. 1426.
Dahal, K., Martyn, G.D., and Vanlerberghe, G.C., Improved photosynthetic performance during severe drought in Nicotiana tabacum overexpressing a nonenergy conserving respiratory electron sink, New Phytol., 2015, vol. 208, p. 382.
Alber, N.A. and Vanlerberghe, G.C., Signaling interactions between mitochondria and chloroplasts in Nicotiana tabacum leaf, Physiol. Plant., 2019, vol. 167, p. 188.
Florez-Sarasa, I., Welchen, E., Racca, S., Gonzalez, D.H., Vallarino, J.G., Fernie, A.R., Ribas-Carbo, M., and Del-Saz, N.F., Cytochrome c deficiency differentially affects the in vivo mitochondrial electron partitioning and primary metabolism depending on the photoperiod, Plants, 2021, vol. 10, p. 444. https://doi.org/10.3390/plants10030444
Albury, M.S., Elliott, C., and Moore, A.L., Towards a structural elucidation of the alternative oxidase in plants, Physiol. Plant., 2009, vol. 137, p. 316.
Tanudji, M., Sjöling, S., Glaser, E., and Whelan, J., Signals required for the import and processing of the alternative oxidase into mitochondria, J. Biol. Chem., 1999, vol. 274 P. 1286.
Selinski, J., Hartmann, A., Kordes, A., Deckers-Hebestreit, G., Whelan, J., and Scheibe, R., Analysis of posttranslational activation of alternative oxidase isoforms, Plant Physiol., 2017, vol. 174, p. 2113.
Guy, R.D. and Vanlerberghe, G.C., Partitioning of respiratory electrons in the dark in leaves of transgenic tobacco with modified levels of alternative oxidase, Physiol. Plant., 2005, vol. 125, p. 171.
Florez-Sarasa, I., Flexas, J., Rasmusson, A.G., Umbach, A.L., Siedow, J.N., and Ribas-Carbo, M., In vivo cytochrome and alternative pathway respiration in leaves of Arabidopsis thaliana plants with altered alternative oxidase under different light conditions, Plant Cell Environ., 2011, vol. 34, p. 1373.
Priault, P., Vidal, G., De Paepe, R., and Ribas-Carbo, M., Leaf age-related changes in respiratory pathways are dependent on complex I activity in Nicotiana sylvestris, Physiol. Plant., 2007, vol. 129.
ACKNOWLEDGMENTS
I thank Prof., Dr. Sci. (Biol.) T.K. Golovko, Cand. Sci. (Biol.) I.O. Velegzhaninov, and anonymous reviewers for careful reading of the manuscript and helpful remarks.
Funding
This work was supported by the Russian Foundation for Basic Research, project no. 20-14-50366 Expansion.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests. The author declares that she has no conflicts of interest.
Statement on the welfare of humans or animals. This article does not contain any studies involving humans or animals performed by the author.
Additional information
Translated by N. Balakshina
Abbreviations: AOX—alternative oxidase; AP—alternative pathway; MRR—mitochondrial retrograde regulation; MFA—monofluoroacetate; PCD—programmed cell death; HSR—hypersensitivity reaction; SA—salicylic acid; TSS—transcription start (initiation) site; TF—transcription factor; chlETC—chloroplast ETC; CP—cytochrome pathway; ER—endoplasmic reticulum; ETC—mitochondrial ETC; АОХ—AOX genes; CARE(s)—cis-acting regulatory element(s); PTP—permeability transition pore; SNP—single-nucleotide polymorphism; UCP—uncoupling proteins; UQ—ubiquinone; 3'-UTR—three prime untranslated region.
Supplementary Information
Rights and permissions
Open Access. This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Garmash, E.V. Signal Pathways for Regulation of Plant Alternative Oxidase Genes’ Expression. Russ J Plant Physiol 69, 1 (2022). https://doi.org/10.1134/S1021443722010058
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1134/S1021443722010058