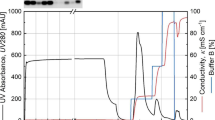
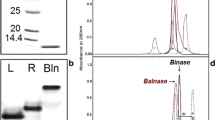
Abstract
Bacillus pumilus ribonuclease (binase) exhibits cytotoxic and oncolytic properties, while causing genotoxic effects at high concentrations. Mutants that have reduced catalytic activity and preserve the antitumor properties of the native enzyme could exert lower toxic side effects. Mutant binase forms with the Lys26Ala and His101Glu single substitutions were obtained by site-directed mutagenesis. A comparative analysis of Escherichia coli- and Bacillus subtilis-based expression systems demonstrated that the latter is better to use to produce the binase mutants. The binase mutants with reduced catalytic activity were isolated and purified to homogeneity by ion exchange chromatography; the maximum yield was 25 mg/L. Catalytic activities of the mutants toward natural RNA-substrates in comparison with those for native binase were estimated at 11% and 0.02%, respectively. Like native binase, the Lys26Ala mutant was found to be cytotoxic to the A549, BT-20, and HuTu 80 tumor cell lines, but did not substantially affect normal WI-38 cells. The His101Glu mutant did not show cytotoxicity.




Similar content being viewed by others
REFERENCES
Ulyanova V., Nadyrova A., Dudkina E., Kuznetsova A., Ahmetgalieva A., Faizullin D., Surchenko Y., Novopashina D., Zuev Y., Kuznetsov N., Ilinskaya O. 2022. Structural and functional differences between homologous bacterial ribonucleases. Int. J. Mol. Sci. 23, 1867.
Ilinskaya O.N., Shah Mahmud R. 2014. Ribonucleases as antiviral agents. Mol. Biol. (Moscow). 48, 616–623.
Makarov A.A., Ilinskaya O.N. 2003. Cytotoxic ribonucleases: Molecular weapons and their targets. FEBS Lett. 540, 15–20.
Ardelt B., Ardelt W., Pozarowski P., Kunicki J., Shogen K., Darzynkiewicz Z. 2007. Cytostatic and cytotoxic properties of Amphinase: A novel cytotoxic ribonuclease from Rana pipiens oocytes. Cell Cycle. 6, 3097‒3102.
Makarov A.A., Kolchinsky A., Ilinskaya O.N. 2008. Binase and other microbial RNases as potential anticancer agents. BioEssays. 30, 781–790.
Mitkevich V.A., Makarov A.A., Ilinskaya O.N. 2014. Cell targets of antitumor ribonucleases. Mol. Biol. (Moscow). 48, 181–188.
Roiz L., Smirnoff P., Bar-Eli M., Schwartz B., Shoseyov O. 2006. ACTIBIND, an actin-binding fungal T-2‑RNase with antiangiogenic and anticarcinogenic characteristics. Cancer. 106, 2295–2308.
Leland P.A., Schultz L.W., Kim B.M., Raines R.T. 1998. Ribonuclease A variants with potent cytotoxic activity. Proc. Natl. Acad. Sci. U. S. A. 95, 10407–10412.
Ilinskaya O.N., Vamvakas S. 1997. Nephrotoxic effects of bacterial ribonucleases in the isolated perfused rat kidney. Toxicology. 120, 55–63.
Yakovlev G.I., Moiseyev G.P., Struminskaya N.K., Borzykh O.A., Kipenskaya L.V., Znamenskaya L.V., Leschinskaya I.B., Chernokalskaya E.B., Hartley R.W. 1994. Mutational analysis of the active site of RNase of Bacillus intermedius (BINASE). FEBS Lett. 354, 305–306.
Rosano, G.L., Ceccarelli E.A. 2014. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 5, 172.
Yoon S.H., Kim S.K., Kim J.F. 2010. Secretory production of recombinant proteins in Escherichia coli. Recent Pat. Biotechnol. 4, 23–29.
Pohl S., Bhavsar G., Hulme J., Bloor A.E., Misirli G., Leckenby M.W., Radford D.S., Smith W., Wipat A., Williamson E.D., Harwood C.R., Cranenburgh R.M. 2013. Proteomic analysis of Bacillus subtilis strains engineered for improved production of heterologous proteins. Proteomics. 13, 3298–3308.
Hartley R.W., Rogerson D.L., Jr., Smeaton J.R. 1972. Production and purification of the extracellular ribonuclease of Bacillus amyloliquefaciens (barnase) and its intracellular inhibitor (barstar). II. Barstar. Prep. Biochem. 2, 243‒250.
Herzberg C., Weidinger L.A.F., Dörrbecker B., Hübner S., Stülke J., Commichau F.M. 2007. SPINE: A method for the rapid detection and analysis of protein–protein interactions in vivo. Proteomics. 7, 4032–4035.
Sambrook J., Russell D.W. 2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Lab. Press. 3rd ed.
Harwood C.R., Cutting S.M. 1991. Molecular Biological Methods for Bacillus. Chichester: Wiley.
Kim R. 2011. Native agarose gel electrophoresis of multiprotein complexes. Cold Spring Harb. Protoc. 2011, 884–887.
Dudkina E., Ulyanova V., Shah Mahmud R., Khodzhaeva V., Dao L., Vershinina V., Kolpakov A., Ilinskaya O. 2016. Three-step procedure for preparation of pure Bacillus altitudinis ribonuclease. FEBS Open Biol. 6, 24–32.
Li W., Zhou X., Lu P. 2004. Bottlenecks in the expression and secretion of heterologous proteins in Bacillus subtilis. Res. Microbiol. 155, 605–610.
Ulyanova V.V., Khodzhaeva V.S., Dudkina E.V., Laykov A.V., Vershinina V.I., Ilinskaya O.N. 2015. Preparations of Bacillus pumilus secreted RNase: One enzyme or two? Microbiology (Moscow). 84, 491–497.
Makarov A.A., Protasevich I.I., Kuznetsova N.V., Fedorov B.B., Korolev S.V., Struminskaya N.K., Bazhulina N.P., Leshchinskaya I.B., Hartley R.W., Kirpichnikov M.P., Yakovlev G.I., Esipova N.G. 1993. Comparative study of thermostability and structure of close homologues—barnase and binase. J. Biomol. Struct. Dyn. 10, 1047–1065.
Golubenko I., Balaban N., Leschinskaya I., Volkova T., Kleyner G., Chepurnova N., Afanasenko G., Dudkin S. 1979. Bacillus intermedius ribonuclease 7P. Purification by chromatography on phosphocellulose and some characteristics of the homogeneous enzyme. Biokhimiya. 44 (4), 640–648.
Shulga A.A., Okorokov A.L., Panov K.I., Kurbanov F.T., Chernov B.K., Skryabin K.G., Kirpichnikov M.P. 1994. Superproduction of Bacillus intermedius 7P ribonuclease (binase) in E. coli. Mol. Biol. 28 (2), 453–463.
Kaur J., Kumar A., Kaur J. 2018. Strategies for optimization of heterologous protein expression in E. coli: Roadblocks and reinforcements. Int. J. Biol. Macromol. 106, 803–822.
Bhatwa A., Wang W., Hassan Y.I., Abraham N., Li X.Z., Zhou T. 2021. Challenges associated with the formation of recombinant protein inclusion bodies in Escherichia coli and strategies to address them for industrial applications. Front. Bioeng. Biotechnol. 9, 630551.
Zhang K., Su L., Wu J. 2020. Recent advances in recombinant protein production by Bacillus subtilis. Annu. Rev. Food Sci. Technol. 11, 295–318.
Okorokov A.L., Panov K.I., Kolbanovskaya E.Y., Karpei-sky M.Y., Polyakov K.M., Wilkinson A.J., Dodson G.G. 1996. Site-directed mutagenesis of the base recognition loop of ribonuclease from Bacillus intermedius (binase). FEBS Lett. 384, 143–146.
Okorokov A.L., Panov K.I., Offen W.A., Mukhortov V.G., Antson A.A., Karpeisky M.Ya., Wilkinson A.J., Dodson G.G. 1997. RNA cleavage without hydrolysis. Splitting the catalytic activities of binase with Asn101 and Thr101 mutations. Protein Eng. 10, 273–278.
Yoshida H. 2001. The ribonuclease T1 family. Methods Enzymol. 341, 28–41.
Bachinsky A.G. 1976. Structure and noise immunity of the genetic code. Zh. Obshch. Biol. 37, 163–173.
Sneath P.H. 1966. Relations between chemical structure and biological activity in peptides. J. Theor. Biol. 12, 157–195.
Castro J., Ribó M., Vilanova M., Benito A. 2021. Strengths and challenges of secretory ribonucleases as antitumor agents. Pharmaceutics. 13, 82.
Gotte G., Menegazzi M. 2019. Biological activities of secretory RNases: Focus on their oligomerization to design antitumor drugs. Front. Immunol. 10, 2626.
Dudkina E.V., Ulyanova V.V., Ilinskaya O.N. 2020. Supramolecular organization as a factor of ribonuclease cytotoxicity. Acta Nat. 12, 24–33.
Ilinskaya O., Singh I., Dudkina E., Ulyanova V., Kayumov A., Barreto G. 2016. Direct inhibition of oncogenic KRAS by Bacillus pumilus ribonuclease (binase). Biochim. Biophys. Acta, Mol. 1863, 1559–1567.
Hollestelle A., Elstrodt F., Nagel J., Kallemeijn W., Schutte M. 2007. Phosphatidylinositol-3-OH kinase or RAS pathway mutations in human breast cancer cell lines. Mol. Cancer Res. 5, 195–201.
Medico E., Russo M., Picco G., Cancelliere C., Valtorta E., Corti G., Buscarino M., Isella M., Lamba S., Martinoglio B., Veronese S., Siena S., Sartore-Bianchi A., Beccuti M., Mottolese M., Linnebacher M., Cordero F., Di Nicolantonio F., Bardelli A. 2015. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat. Commun. 6, 7002.
Chen Y. H., Lv H., Shen N., Wang X. M., Tang S., Xiong B., Ding J., Geng M., Huang, M. 2020. EPHA2 feedback activation limits the response to PDEδ inhibition in KRAS-dependent cancer cells. Acta Pharmacol. Sin. 41, 270–277.
Ulyanova V., Dudkina E., Nadyrova A., Kalashnikov V., Surchenko Y., Ilinskaya O. 2020. The cytotoxicity of RNase-derived peptides. Biomolecules. 11, 16.
Zelenikhin P.V., Ead Mokhamed I.S., Nadyrova A.I., Sirotkina A.A., Ulyanova V.V., Mironova N.L., Mitkevich V.A., Makarov A.A., Zenkova M.A., Ilinskaya O.N. 2020. Bacillus pumilus ribonuclease inhibits migration of human duodenum adenocarcinoma HuTu 80 cells. Mol. Biol. (Moscow). 54, 128–133. https://doi.org/10.1134/S0026893320010173
Alford S.C., Pearson J.D., Carette A., Ingham R.J., Howard P.L. 2009. Alpha-sarcin catalytic activity is not required for cytotoxicity. BMC Biochem. 10, 9.
Navarro S., Aleu J., Jiménez M., Boix E., Cuchillo C.M., Nogués M.V. 2008. The cytotoxicity of eosinophil cationic protein/ribonuclease 3 on eukaryotic cell lines takes place through its aggregation on the cell membrane. Cell. Mol. Life Sci. 65, 324–337.
Ilinskaya O., Ivanchenko O.B., Karamova N.S. 1995. Bacterial ribonuclease: Mutagenic effect in microbial test-systems. Mutagenesis. 10, 165–170.
Ilinskaya O.N., Karamova N.S., Ivanchenko O.B., Kipenskaya L.V. 1996. SOS-inducing ability of native and mutant microbial ribonucleases. Mutat. Res. 354, 203–209.
Funding
This work was supported by the Russian Science Foundation (project no. 21-74-10036) and the Ministry of Science and Higher Education of the Russian Federation (the program “Priority-2030”).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest. The authors declare that they have no conflicts of interest.
This work does not contain any studies involving animals or human subjects performed by any of the authors.
Additional information
Translated by T. Tkacheva
Rights and permissions
About this article
Cite this article
Nadyrova, A.I., Kosnyrev, A.S., Ulyanova, V.V. et al. Efficiency of Escherichia coli and Bacillus subtilis Expression Systems for Production of Binase Mutants. Mol Biol 57, 825–835 (2023). https://doi.org/10.1134/S002689332305014X
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S002689332305014X