Abstract—
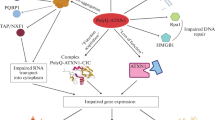
Polyglutamine diseases are rare, inherited neurodegenerative pathologies that arise as a result of expansion of trinucleotide CAG repeats in the coding segment of certain genes. This expansion leads to the appearance of mRNA with abnormally long repetitive CAG triplets (mCAG-RNA) and proteins with polyglutamine (PolyQ) tracts in the cells, which is why these pathologies are commonly termed polyglutamine diseases, or PolyQ diseases. To date, nine PolyQ diseases have been described: Huntington’s disease, dentatorubral pallidoluysian atrophy (DRPLA), spinal and bulbar muscular atrophy (SBMA), and six different types of spinocerebellar ataxia (SCA 1, 2, 3, 6, 7, and 17). PolyQ diseases lead to serious, constantly progressing dysfunctions of the nervous and/or muscular systems, and there currently exists no efficacious therapy for any of them. Recent studies have convincingly shown that mCAG-RNA can actively participate in the pathological process during the development of PolyQ diseases. Mutant RNA is involved in a wide range of molecular mechanisms, ultimately leading to disruption of the functions of transcription, splicing, translation, cytosol structure, RNA transport from the nucleus to the cytoplasm, and, finally, to neurodegeneration. This review discusses the involvement of mutant mCAG-RNA in neurodegenerative processes in PolyQ diseases.

Similar content being viewed by others
REFERENCES
Aronin N., DiFiglia M. 2014. Huntingtin-lowering strategies in Huntington’s disease: Antisense oligonucleotides, small RNAs, and gene editing. Movement Disorders.29, 1455‒1461.
Hu J., Matsui M., Gagnon K.T., Schwartz J.C., Gabillet S., Arar K., Wu J., Bezprozvanny I., Corey D.R. 2009. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat. Biotechnol.27, 478.
Hu J., Liu J., Corey D.R. 2010. Allele-selective inhibition of huntingtin expression by switching to an miRNA-like RNAi mechanism. Chem. Biol.17, 1183‒1188.
Sharp A.H., Loev S.J., Schilling G., Li S.H., Li X.J., Bao J., Wagster M.V., Kotzuk J.A., Steiner J.P., Lo A., Hedreen J. 1995. Widespread expression of Huntington’s disease gene (IT15) protein product. Neuron. 14, 1065‒1074.
Illarioshkin S.N., Klyushnikov S.A., Seliverstov Yu.A. 2018. Bolezn’ Gentingtona (Huntington’s Disease). Moscow: Atmosfera.
Nasir J., Floresco S.B., O’Kusky J.R., Diewert V.M., Richman J.M., Zeisler J., Borowski A., Marth J.D., Phillips A.G., Hayden M.R. 1995. Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 81, 811‒823.
Wexler N.S., Young A.B., Tanzi R.E., Travers H., Starosta-Rubinstein S., Penney J.B., Snodgrass S.R., Shoulson I., Gomez F., Ramos Arroyo M.A., Penchaszadeh G.K. 1987. Homozygotes for Huntington’s disease. Nature. 326, 194.
Myers R.H., Leavitt J.L., Farrer L.A., Jagadeesh J., McFarlane H., Mastromauro C.A., Mark R.J., Gusella J.F. 1989. Homozygote for Huntington disease. Am. J. Hum. Genet.45, 615.
Evers M.M., Schut M.H., Pepers B.A., Atalar M., van Belzen M.J., Faull R.L., Roos R.A., van Roon-Mom W.M. 2015. Making (anti-)sense out of huntingtin levels in Huntington disease. Mol. Neurodegener.10, 21.
Shin A., Shin B., Shin J.W., Kim K.H., Atwal R.S., Hope J.M., Gillis T., Leszyk J.D., Shaffer S.A., Lee R., Kwak S., MacDonald M.E., Gusella J.F., Seong I.S., Lee J.M. 2017. Novel allele-specific quantification methods reveal no effects of adult onset CAG repeats on HTT mRNA and protein levels. Hum. Mol. Genet.26, 1258‒1267.
Neueder A., Landles C., Ghosh R., Howland D., Myers R.H., Faull R.L., Tabrizi S.J., Bates G.P. 2017. The pathogenic exon 1 HTT protein is produced by incomplete splicing in Huntington’s disease patients. Sci. Rept.7, 1307.
Sathasivam K., Neueder A., Gipson T.A., Landles C., Benjamin A.C., Bondulich M.K., Smith D.L., Faull R.L., Roos R.A., Howland D., Detloff P.J., Housman D.E., Bates G.P. 2013. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl. Acad. Sci. U. S. A.110, 2366‒2370.
Neueder A., Dumas A.A., Benjamin A.C., Bates G.P. 2018. Regulatory mechanisms of incomplete huntingtin mRNA splicing. Nat. Commun.9, 3955.
Romo L., Ashar-Patel A., Pfister E., Aronin N. 2017. Alterations in mRNA 3′ UTR isoform abundance accompany gene expression changes in human Huntington’s disease brains. Cell Rept.20, 3057‒3070.
Romo L., Mohn E.S., Aronin N. 2018. A fresh look at huntingtin mRNA processing in Huntington’s disease. J. Huntington’s Dis.7, 101‒108.
Xu H., An J.J., Xu B. 2017. Distinct cellular toxicity of two mutant huntingtin mRNA variants due to translation regulation. PLoS One.12, e0177610.
Khalil A.M., Faghihi M.A., Modarresi F., Brothers S.P., Wahlestedt C. 2008. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS One.3, e1486.
Daughters R.S., Tuttle D.L., Gao W., Ikeda Y., Moseley M.L., Ebner T.J., Swanson M.S., Ranum L.P. 2009. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genetics. 5, e1000600.
Chung D.W., Rudnicki D.D., Yu L., Margolis R.L. 2011. A natural antisense transcript at the Huntington’s disease repeat locus regulates HTT expression. Hum. Mol. Genet.20, 3467‒3477.
Li P.P., Sun X., Xia G., Arbez N., Paul S., Zhu S., Peng H.B., Ross C.A., Koeppen A.H., Margolis R.L., Pulst S.M., Ashizawa T., Rudnicki D.D. 2016. ATXN2-AS, a gene antisense to ATXN2, is associated with spinocerebellar ataxia type 2 and amyotrophic lateral sclerosis. Ann. Neurol.80, 600‒615.
de Mezer M., Wojciechowska M., Napierala M., Sobczak K., Krzyzosiak W.J. 2011. Mutant CAG repeats of huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res.39, 3852‒3863.
Wojciechowska M., Krzyzosiak W.J. 2011. Cellular toxicity of expanded RNA repeats: Focus on RNA foci. Hum. Mol. Genet.20, 3811‒3821.
Urbanek M.O., Krzyzosiak W.J. 2016. RNA FISH for detecting expanded repeats in human diseases. Methods.98, 115‒123.
Taneja K.L., McCurrach M., Schalling M., Housman D., Singer R.H. 1995. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell. Biol.128, 995‒1002.
Wheeler T.M., Thornton C.A. 2007. Myotonic dystrophy: RNA-mediated muscle disease. Curr. Opin. Neurol.20, 572‒576.
Mooers B.H., Logue J.S., Berglund J.A. 2005. The structural basis of myotonic dystrophy from the crystal structure of CUG repeats. Proc. Natl. Acad. Sci. U. S. A.102, 16626‒16631.
Yuan Y., Compton S.A., Sobczak K., Stenberg M.G., Thornton C.A., Griffith J.D., Swanson M.S. 2007. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res.35, 5474‒5486.
Nakamori M., Sobczak K., Puwanant A., Welle S., Eichinger K., Pandya S., Dekdebrun J., Heatwole C.R., McDermott M.P., Chen T., Cline M., Tawil R., Osborne R.J., Wheeler T.M., Swanson M.S., et al. 2013. Splicing biomarkers of disease severity in myotonic dystrophy. Ann. Neurol.74, 862‒872.
Botta A., Vallo L., Rinaldi F., Bonifazi E., Amati F., Biancolella M., Gambardella S., Mancinelli E., Angelini C., Meola G., Novelli G. 2006. Gene expression analysis in myotonic dystrophy: Indications for a common molecular pathogenic pathway in DM1 and DM2. Gene expression, J. Liver Res.13, 339‒351.
Salvatori S., Furlan S., Fanin M., Picard A., Pastorello E., Romeo V., Trevisan C.P., Angelini C. 2009. Comparative transcriptional and biochemical studies in muscle of myotonic dystrophies (DM1 and DM2). Neurol. Sci.30, 185‒192.
Querido E., Gallardo F., Beaudoin M., Ménard C., Chartrand P. 2011. Stochastic and reversible aggregation of mRNA with expanded CUG-triplet repeats. J. Cell. Sci.124, 1703‒1714.
Konieczny P., Stepniak-Konieczna E., Sobczak K. 2014. MBNL proteins and their target RNAs, interaction and splicing regulation. Nucleic Acids Res.42, 10873‒10887.
Sobczak K., Wheeler T.M., Wang W., Thornton C.A. 2013. RNA interference targeting CUG repeats in a mouse model of myotonic dystrophy. Mol. Therapy.21, 380‒387.
Jauvin D., Chrétien J., Pandey S. K., Martineau L., Revillod L., Bassez G., Thornton C.A. 2017. Targeting DMPK with antisense oligonucleotide improves muscle strength in myotonic dystrophy type 1 mice. Mol. Therapy–Nucl. Acids.7, 465‒474.
Rzuczek S.G., Colgan L.A., Nakai Y., Cameron M.D., Furling D., Yasuda R., Disney M.D. 2017. Precise small-molecule recognition of a toxic CUG RNA repeat expansion. Nat. Chem. Biol.13, 188.
Angelbello A.J., Rzuczek S.G., Mckee K.K., Chen J.L., Olafson H., Cameron M.D., Moss W.N., Wang E.T., Disney M.D. 2019. Precise small-molecule cleavage of an r (CUG) repeat expansion in a myotonic dystrophy mouse model. Proc. Natl. Acad. Sci. U. S. A.116, 7799–7804.
Yuan Y., Compton S.A., Sobczak K., Stenberg M.G., Thornton C.A., Griffith J.D., Swanson M.S. 2007. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res.35, 5474‒5486.
Tawani A., Kumar A. 2015. Structural insights reveal the dynamics of the repeating r (CAG) transcript found in Huntington’s disease (HD) and spinocerebellar ataxias (SCAs). PLoS One.10, e0131788.
Sobczak K., Michlewski G., de Mezer M., Kierzek E., Krol J., Olejniczak M., Kierzek R., Krzyzosiak W.J. 2010. Structural diversity of triplet repeat RNAs. J. Biol. Chem.285, 12755‒12764.
Urbanek M.O., Jazurek M., Switonski P.M., Figura G., Krzyzosiak W.J. 2016. Nuclear speckles are detention centers for transcripts containing expanded CAG repeats. Biochim. Biophys. Acta—Mol.Basis Disease.1862, 1513‒1520.
Mykowska A., Sobczak K., Wojciechowska M., Kozlowski P., Krzyzosiak W.J. 2011. CAG repeats mimic CUG repeats in the misregulation of alternative splicing. Nucleic Acids Res.39, 8938‒8951.
Mizielinska S., Grönke S., Niccoli T., Ridler C.E., Clayton E.L., Devoy A., Moens T., Norona F.E., Woollacott I.O.C., Pietrzyk J., Cleverley K., Nicoll A.J., Pickering-Brown S., Dols J., Cabecinha M., et al. 2014. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science.345, 1192‒1194.
Hodges A., Strand A.D., Aragaki A.K., Kuhn A., Sengstag T., Hughes G., Elliston L.A., Hartog C., Goldstein D.R., Thu D., Hollingsworth Z.R., Collin F., Synek B., Holmans P.A., Young A.B., et al. 2006. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum. Mol. Genet.15, 965‒977.
Krol J., Fiszer A., Mykowska A., Sobczak K., de Mezer M., Krzyzosiak W.J. 2007. Ribonuclease dicer cleaves triplet repeat hairpins into shorter repeats that silence specific targets. Mol. Cell.25, 575‒586.
Bañez-Coronel M., Porta S., Kagerbauer B., Mateu-Huertas E., Pantano L., Ferrer I., Guzmán M., Estivill X., Martí E. 2012. A pathogenic mechanism in Huntington’s disease involves small CAG-repeated RNAs with neurotoxic activity. PLoS Genet.8, e1002481.
Reinhardt A., Feuillette S., Cassar M., Callens C., Thomassin-Bourrel H., Birman S., Lecourtois M., Antoniewski C., Tricoire H. 2012. Lack of miRNA misregulation at early pathological stages in Drosophila neurodegenerative disease models. Front. Genet.3, 226.
Murmann A.E., Gao Q.Q., Putzbach W.E., Patel M., Bartom E.T., Law C.Y., Bridgeman B., Chen S., McMahon K.M., Thaxton C.S., Peter M.E. 2018. Small interfering RNAs based on huntingtin trinucleotide repeats are highly toxic to cancer cells. EMBO Repts.19, e45336.
Sørensen S.A., Fenger K., Olsen J.H. 1999. Significantly lower incidence of cancer among patients with Huntington disease: An apoptotic effect of an expanded polyglutamine tract? Cancer.86, 1342‒1346.
Schilling J., Broemer M., Atanassov I., Duernberger Y., Vorberg I., Dieterich C., Dagane A., Dittmar G., Wanker E., van Roon-Mom W., Winter J., Krauß S. 2019. Deregulated splicing is a major mechanism of RNA-induced toxicity in Huntington’s disease. J. Mol. Biol. 19, 1869‒1877.
Lee J., Hwang Y.J., Boo J.H., Han D., Kwon O.K., Todorova K., Kowall N.W., Kim Y., Ryu H. 2011. Dysregulation of upstream binding factor-1 acetylation at K352 is linked to impaired ribosomal DNA transcription in Huntington’s disease. Cell Death Differ.18, 1726.
Tsoi H., Lau T.C.K., Tsang S.Y., Lau K.F., Chan H.Y.E. 2012. CAG expansion induces nucleolar stress in polyglutamine diseases. Proc. Natl. Acad. Sci. U. S. A.109, 13428‒13433.
Zhang Q., Chen Z.S., An Y., Liu H., Hou Y., Li W., Lau K.F., Koon A.C., Ngo J.C.K., Chan H. Y.E. 2018. A peptidylic inhibitor for neutralizing expanded CAG RNA-induced nucleolar stress in polyglutamine diseases. RNA.24, 486‒498.
Bañez-Coronel M., Ayhan F., Tarabochia A.D., Zu T., Perez B.A., Tusi S.K., Pletnikova O., Borchelt D.R., Ross C.A., Margolis R.L., Yachnis A.T., Troncoso J.C., Ranum L.P. 2015. RAN translation in Huntington disease. Neuron.88, 667‒677.
Volovikov E.A., Davidenko A.V., Lagar’kova M.A. 2019. Molecular mechanisms of type 1 ataxia. Russ. J. Genet.55 (in press).
Krauss S., Griesche N., Jastrzebska E., Chen C., Rutschow D., Achmüller C., Dorn S., Boesch S.M., Lalowski M., Wanker E., Schneider R., Schweiger S. 2013. Translation of HTT mRNA with expanded CAG repeats is regulated by the MID1-PP2A protein complex. Nat. Commun.4, 1511.
Griesche N., Schilling J., Weber S., Rohm M., Pesch V., Matthes F., Auburger G., Krauss S. 2016. Regulation of mRNA translation by MID1: A common mechanism of expanded CAG repeat RNAs. Front. Cell Neurosci.7, 226.
La Spada A.R., Taylor J.P. 2010. Repeat expansion disease: Progress and puzzles in disease pathogenesis. Nat. Rev. Genet.11, 247.
Babić Leko M., Župunski V., Kirincich J., Smilović D., Hortobágyi T., Hof P.R., Šimić G. 2019. Molecular mechanisms of neurodegeneration related to C9orf72 hexanucleotide repeat expansion. Behav. Neurol.2019, 2909168
Lee D.Y., McMurray C.T. 2014. Trinucleotide expansion in disease: Why is there a length threshold? Curr. Opin. Genet. Dev.26, 131‒140.
Jain A., Vale R.D. 2017. RNA phase transitions in repeat expansion disorders. Nature. 546, 243.
Fay M.M., Anderson P.J. 2018. The role of RNA in biological phase separations. J. Mol. Biol.430, 4685–4701.
Boeynaems S., Alberti S., Fawzi N.L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch L., Tompa P., Fuxreiter M. 2018. Protein phase separation: A new phase in cell biology. Trends Cell Biol.28, 420‒435.
Sabari B.R., Dall’Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K., Abraham B.J., Hannett N.M., Zamudio A.V., Manteiga J.C., Li C.H., Guo Y.E., Day D.S., Schuijers J., Vasile E., et al. 2018. Coactivator condensation at super-enhancers links phase separation and gene control. Science.361, eaar3958.
Larson A.G., Narlikar G.J. 2018. The role of phase separation in heterochromatin formation, function, and regulation. Biochemistry.57, 2540‒2548.
Singatulina A.S., Hamon L., Sukhanova M.V., Desforges B., Joshi V., Bouhss A., Lavrik O.I., Pastré D. 2019. PARP-1 activation directs FUS to DNA damage sites to form PARG-reversible compartments enriched in damaged DNA. Cell Rept.27, 1809‒1821.
Kato M., Yang Y.S., Sutter B.M., Wang Y., McKnight S.L., Tu B.P. 2019. Redox state controls phase separation of the yeast ataxin-2 protein via reversible oxidation of its methionine-rich low-complexity domain. Cell.177, 711‒721.
Harper S.Q., Staber P.D., He X., Eliason S.L., Martins I.H., Mao Q., Yang L, Kotin R.M., Paulson H.L., Davidson B. L. 2005. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc. Natl. Acad. Sci. U. S. A.102, 5820‒5825.
http://www.uniqure.com/gene-therapy/huntingtons-disease.php
Bennett C.F., Swayze E.E. 2010. RNA targeting therapeutics: Molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol.50, 259‒293.
https://clinicaltrials.gov/ct2/show/NCT02519036
Naryshkin N.A., Weetall M., Dakka A., Narasimhan J., Zhao X., Feng Z., Ling K.K., Karp G.M., Qi H., Woll M.G., Chen G., Zhang N., Gabbeta V., Vazirani P., Bhattacharyya A., et al. 2014. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science.345, 688‒693.
Doherty E.M. 2017. Screening approaches to identify small-molecule modulators of huntingtin protein levels. Abstr. CHDI Foundation Annual Therapeutics Conference, Malta, 2017.
Rousseaux M.W., Tschumperlin T., Lu H.C., Lackey E.P., Bondar V.V., Wan Y.W., Tan Q., Adamski C.J., Friedrich J., Twaroski K., Chen W., Tolar J., Henzler C., Sharma A., Bajić A., et al. 2018. ATXN1-CIC complex is the primary driver of cerebellar pathology in spinocerebellar ataxia type 1 through a gain-of-function mechanism. Neuron.97, 1235‒1243.
Takahashi K., Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell.126, 663‒676.
Nekrasov E.D., Vigont V.A., Klyushnikov S.A., Lebedeva O.S., Vassina E.M., Bogomazova A.N., Chestkov I.V., Semashko T.A., Kiseleva E., Suldina L.A., Bobrovsky P.A., Zimina O.A., Ryazantseva M.A., Skopin A.Y., Illarioshkin S.N., et al. 2016. Manifestation of Huntington’s disease pathology in human induced pluripotent stem cell-derived neurons. Mol. Neurodegeneration.11, 27.
Funding
This work was supported by the Russian Science Foundation (project no. 19-15-00425).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain studies performed by the authors involving animals or human subjects.
Additional information
Translated by D. Timchenko
Abbreviations: GABA, gamma-aminobutyric acid; IPSC, induced pluripotent stem cells; ASO, antisense oligonucleotide; DM, myotonic dystrophy; DMPK, myotonic dystrophy protein kinase; DRPA, dentatorubral-pallidoluysian atrophy; FDA, Food and Drug Administration; FISH, fluorescence in situ hybridization; HTT, huntingtin protein; MBNL1, muscleblindlike splicing regulator 1; mCAG-RNA, mRNA with CAG repeat expansion; mCUG-RNA, mRNA with CUG expansion; mHTT, mutant huntingtin; MID1 (Midline 1), E3 ubiquitin-protein ligase Midline-1; OMIM, Online Mendelian Inheritance in Man; PolyQ, polyglutamine; PP2A, protein phosphatase 2A; RAN, repeat associated non-ATG; SBMA, spinal and bulbar muscular atrophy; SCA, spinocerebellar ataxia; sCAG, 21 bp-long RNA duplexes composed of CAG repeats.
Rights and permissions
About this article
Cite this article
Bogomazova, A.N., Eremeev, A.V., Pozmogova, G.E. et al. The Role of Mutant RNA in the Pathogenesis of Huntington’s Disease and Other Polyglutamine Diseases. Mol Biol 53, 838–849 (2019). https://doi.org/10.1134/S0026893319060037
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893319060037