Abstract
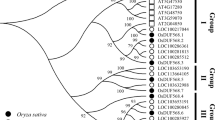
With the advance of sequencing technology, the number of sequenced plant genomes has been rapidly increasing. However, understanding of the gene function in these sequenced genomes lags far behind; as a result, many coding plant sequences in public databases are annotated as proteins with domains of unknown function (DUF). Function of a protein family DUF810 in rice is not known. In this study, we analysed seven members of OsDU810 (OsDUF810.1–OsDUF810.7) family with three distinct motifs in rice Nipponbare. By phylogenetic analysis, OsDUF810 proteins fall into three major groups (I, II, III). Expression patterns of the seven corresponding OsDUF810 protein-encoding genes in 15 different rice tissues vary. Under drought, salt, cold and heat stress conditions and ABA treatment, the expression of OsDUF810.7 significantly increases. Overexpression of this protein in E. coli lead to a significant enhancement of catalase (CAT) and peroxidase (POD) activities, and improved bacterial resistance to salt and drought.
Similar content being viewed by others
Abbreviations
- ABA:
-
abscisic acid
- CAT:
-
catalase
- DUF:
-
the domain of unknown function
- POD:
-
peroxidase
- SOD:
-
superoxide dismutase
- ROS:
-
reactive oxygen species
References
Boyer J.S. 1982. Plant productivity and environment. Science. 218, 443–448.
Bohnert H.J., Gong Q., Li P., Ma S. 2006. Unraveling abiotic stress tolerance mechanisms: Getting genomics going. Curr. Opin. Plant Biol. 9, 180–188.
Serrano R., Rodriguez-Navarro A. 2001. Ion homeostasis during salt stress in plants. Curr. Opin. Cell Biol. 13, 399–404.
Park S.Y., Fung P., Nishimura N., Jensen D.R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T.F., Alfred S.E., Bonetta D., Finkelstein R., Provart N.J., Desveaux D., et al. 2009. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 324, 1068–1071.
Kwak J.M., Mori I.C., Pei Z.M., Leonhardt N., Torres M.A., Dangl J.L., Bloom R.E., Bodde S., Jones J.D., Schroeder J.I. 2003. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22, 2623–2633.
Sutter J.U., Sieben C., Hartel A., Eisenach C., Thiel G., Blatt M.R. 2007. Abscisic acid triggers the endocytosis of the arabidopsis KAT1 K+ channel and its recycling to the plasma membrane. Curr. Biol. 17, 1396–1402.
Fujita Y., Fujita M., Shinozaki K., Yamaguchi-Shinozaki K. 2011. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124, 509–525.
Bateman A., Coggill P., Finn R.D. 2010. DUFs: families in search of function. Acta. Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 66, 1148–1152.
Bischoff V., Nita S., Neumetzler L., Schindelasch D., Urbain A., Eshed R., Persson S., Delmer D., Scheible W.R. 2010. TRICHOME BIREFRINGENCE and its homolog AT5G01360 encode plant-specific DUF231 proteins required for cellulose biosynthesis in Arabidopsis. Plant Physiol. 153, 590–602.
Cao X., Yang K.Z., Xia C., Zhang X.Q., Chen L.Q., Ye D. 2010. Characterization of DUF724 gene family in Arabidopsis thaliana. Plant Mol. Biol. 72, 61–73.
Jones-Rhoades M.W., Borevitz J.O., Preuss D. 2007. Genome-wide expression profiling of the Arabidopsis female gametophyte identifies families of small, secreted proteins. PLoS Genet. 3, 1848–1861.
He X., Hou X., Shen Y., Huang Z. 2011. TaSRG, a wheat transcription factor, significantly affects salt tolerance in transgenic rice and Arabidopsis. FEBS Lett. 585, 1231–1237.
Kim S.J., Ryu M.Y., Kim W.T. 2012. Suppression of Arabidopsis RING-DUF1117 E3 ubiquitin ligases, AtRDUF1 and AtRDUF2, reduces tolerance to ABAmediated drought stress. Biochem. Biophys. Res. Commun. 420, 141–147.
Luo C., Guo C., Wang W., Wang L., Chen L. 2014. Overexpression of a new stress-repressive gene OsDSR2 encoding a protein with a DUF966 domain increases salt and simulated drought stress sensitivities and reduces ABA sensitivity in rice. Plant Cell Rep. 33, 323–336.
Wang L., Shen R., Chen L.T., Liu Y.G. 2014. Characterization of a novel DUF1618 gene family in rice. J. Integr. Plant Biol. 56, 151–158.
Guo C., Luo C., Guo L., Li M., Guo X., Zhang Y., Wang L., Chen L. 2016. OsSIDP366, a DUF1644 gene, positively regulates responses to drought and salt stresses in rice. J. Integr. Plant Biol. 58, 492–502.
Letunic I., Doerks T., Bork P. 2014. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 43, 257–260.
Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. 2009. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–208.
Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J.D., Higgins D.G. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539.
Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599.
Li L., Liu C., Lian X. 2010. Gene expression profiles in rice roots under low phosphorus stress. Plant Mol. Biol. 72, 423–432.
Li L., Ye T., Gao X., Xu J., Xie C., Zhu J., Deng X., Wang P., Xu Z. 2017. Molecular characterization and functional analysis of the OsPsbR gene family in rice. Mol. Genet. Genomics. 292, 271–281.
Livak K.J., Schmittgen T.D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25, 402–408.
Bradford M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 72, 248–254.
LaVallie E.R., DiBlasio E.A., Kovacic S., Grant K.L., Schendel P.F., McCoy J.M. 1993. A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Biotechnology. 11, 187–193.
Liang Y., Chen Q., Liu Q., Zhang W., Ding R. 2003. Exogenous silicon (Si) increases antioxidant enzyme activity and reduces lipid peroxidation in roots of saltstressed barley (Hordeum vulgare L.). J. Plant Physiol. 160, 1157–1164.
Wang X., Shi X., Hao B., Ge S., Luo J. 2005. Duplication and DNA segmental loss in the rice genome: Implications for diploidization. New Phytol. 165, 937–946.
Ito Y., Katsura K., Maruyama K., Taji T., Kobayashi M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. 2006. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 47, 141–153.
Dubouzet J.G., Sakuma Y., Ito Y., Kasuga M., Dubouzet E.G., Miura S., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. 2003. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33, 751–763.
Miller G., Shulaev V., Mittler R. 2008. Reactive oxygen signaling and abiotic stress. Physiol. Plant. 133, 481–489.
Author information
Authors and Affiliations
Corresponding author
Additional information
Published in Russian in Molekulyarnaya Biologiya, 2018, Vol. 52, No. 4, pp. 567–575.
The text was submitted by the author(s) in English.
Rights and permissions
About this article
Cite this article
Li, LH., Lv, MM., Li, X. et al. The Rice OsDUF810 Family: OsDUF810.7 May be Involved in the Tolerance to Salt and Drought. Mol Biol 52, 489–496 (2018). https://doi.org/10.1134/S002689331804012X
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S002689331804012X