Abstract
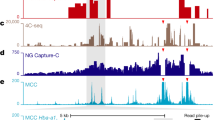
The spatial organization of the eukaryotic genome is closely related to its function. In particular, interactions of gene promoters with distant enhancer elements in active chromatin hubs and gene recruitment to common transcription factories play an important role in regulating gene transcription. Tissue-specific genes are mostly used as models to study the spatial interactions of genomic regulatory elements, while little is known as to what extent the spatial organization of chromosomes is guided by housekeeping genes, which are transcribed in the majority of cells and are considerably more abundant than transcribed tissue-specific genes. To address the issue, chromosome conformation capture on chip (4C) was employed in a genomewide probing of spatial contacts for the chicken housekeeping genes CARHSP1 and TRAP1, which are on chromosome 14. Their promoters showed a higher frequency of interactions with transcriptionally active chromosome regions and regions enriched in Sp1 general transcription factor-binding sites and CpG islands, which both mark the promoters of housekeeping genes. No such preferences were observed for a gene-poor chromosome 14 region. Further evidence for the association of housekeeping gene promoters was obtained in independent cytological visualization of nonmethylated CpG islands in individual human cell nuclei. CpG islands were observed to cluster in the nuclear space. The results testify that the interaction of housekeeping gene promoters is an important factor that determines the spatial organization of interphase chromosomes.
Similar content being viewed by others
Abbreviations
- 3C:
-
chromosome conformation capture
- 4C:
-
chromosome conformation capture on chip
- DAPI:
-
4′,6-diamidino-2-phenylindole
- FISH:
-
fluorescence in situ hybridization
References
Holwerda S., de Laat W. 2012. Chromatin loops, gene positioning, and gene expression. Front. Genet. 3, 217.
de Wit E., de Laat W. 2012. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 26, 11–24.
Palstra R.J. 2009. Close encounters of the 3C kind: long-range chromatin interactions and transcriptional regulation. Brief. Funct. Genomic Proteomic. 8, 297–309.
Gavrilov A.A., Razin S.V., Iarovaia O.V. 2012. C-methods to study 3D organization of the eukaryotic genome. Biopolymers Cell. 28, 245–251.
Palstra R.J., Tolhuis B., Splinter E., Nijmeijer R., Grosveld F., de Laat W. 2003. The beta-globin nuclear compartment in development and erythroid differentiation. Nature Genet. 35, 190–194.
de Laat W., Grosveld F. 2003. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 11, 447–459.
Sutherland H., Bickmore W.A. 2009. Transcription factories: gene expression in unions? Nature Rev. Genet. 10, 457–466.
Razin S.V., Gavrilov A.A., Pichugin A., Lipinski M., Iarovaia O.V., Vassetzky Y.S. 2011. Transcription factories in the context of the nuclear and genome organization. Nucleic Acids Res. 39, 9085–9092.
Papantonis A., Cook P.R. 2013. Transcription factories: Genome organization and gene regulation. Chem. Rev. 113, 8683–8705.
Osborne C.S., Chakalova L., Brown K.E., Carter D., Horton A., Debrand E., Goyenechea B., Mitchell J.A., Lopes S., Reik W., Fraser P. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nature Genet. 36, 1065–1071.
Faro-Trindade I., Cook P.R. 2006. Transcription factories: Structures conserved during differentiation and evolution. Biochem. Soc. Trans. 34, 1133–1137.
Carter D.R., Eskiw C., Cook P.R. 2008. Transcription factories. Biochem. Soc. Trans. 36, 585–589.
Martin S., Failla A.V., Spori U., Cremer C., Pombo A. 2004. Measuring the size of biological nanostructures with spatially modulated illumination microscopy. Mol. Biol. Cell. 15, 2449–2455.
Schoenfelder S., Sexton T., Chakalova L., Cope N.F., Horton A., Andrews S., Kurukuti S., Mitchell J.A., Umlauf D., Dimitrova D.S., Eskiw C.H., Luo Y., Wei C.L., Ruan Y., Bieker J.J., Fraser P. 2010. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nature Genet. 42, 53–61.
Osborne C.S., Chakalova L., Mitchell J.A., Horton A., Wood A.L., Bolland D.J., Corcoran A.E., Fraser P. 2007. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 5, e192.
Philonenko E.S., Klochkov D.B., Borunova V.V., Gavrilov A.A., Razin S.V., Iarovaia O.V. 2009. TMEM8, a non-globin gene entrapped in the globin web. Nucleic Acids Res. 37, 7394–7406.
Zhou G.L., Xin L., Song W., Di L.J., Liu G., Wu X.S., Liu D.P., Liang C.C. 2006. Active chromatin hub of the mouse alpha-globin locus forms in a transcription factory of clustered housekeeping genes. Mol. Cell. Biol. 26, 5096–5105.
Simonis M., Klous P., Splinter E., Moshkin Y., Willemsen R., de Wit E., van Steensel B., de Laat W. 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nature Genet. 38, 1348–1354.
Tolhuis B., Blom M., Kerkhoven R.M., Pagie L., Teunissen H., Nieuwland M., Simonis M., de Laat W., van Lohuizen M., van Steensel B. 2011. Interactions among Polycomb domains are guided by chromosome architecture. PLoS Genet. 7, e1001343.
Splinter E., de Wit E., van de Werken H.J., Klous P., de Laat W. 2012. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: from fixation to computation. Methods. 58, 221–230.
Kundu T.K., Rao M.R. 1999. CpG islands in chromatin organization and gene expression. J. Biochem. 125, 217–222.
Deaton A.M., Bird A. 2011. CpG islands and the regulation of transcription. Genes Dev. 25, 1010–1022.
Beug H., Palmieri S., Freudenstein C., Zentgraf H., Graf T. 1982. Hormone-dependent terminal differentiation in vitro of chicken erythroleukemia cells transformed by ts mutants of avian erythroblastosis virus. Cell. 28, 907–919.
Tolhuis B., Palstra R.J., Splinter E., Grosveld F., de Laat W. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 10, 1453–1465.
Gavrilov A., Eivazova E., Priozhkova I., Lipinski M., Razin S., Vassetzky Y. 2009. Chromosome conformation capture (from 3C to 5C) and its ChIP-based modification. Methods Mol. Biol. 567, 171–188.
Langmead B., Schatz M.C., Lin J., Pop M., Salzberg S.L. 2009. Searching for SNPs with cloud computing. Genome Biol. 10, R134.
Gardiner-Garden M., Frommer M. 1987. CpG islands in vertebrate genomes. J. Mol. Biol. 196, 261–282.
Splinter E., de Wit E., Nora E.P., Klous P., van de Werken H.J., Zhu Y., Kaaij L.J., van Ijcken W., Gribnau J., Heard E., de Laat W. 2011. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 25, 1371–1383.
Groblewski G.E., Yoshida M., Bragado M.J., Ernst S.A., Leykam J., Williams J.A. 1998. Purification and characterization of a novel physiological substrate for calcineurin in mammalian cells. J. Biol. Chem. 273, 22738–22744.
Felts S.J., Owen B.A., Nguyen P., Trepel J., Donner D.B., Toft D.O. 2000. The hsp90-related protein TRAP1 is a mitochondrial protein with distinct functional properties. J. Biol. Chem. 275, 3305–3312.
Song H.Y., Dunbar J.D., Zhang Y.X., Guo D., Donner D.B. 1995. Identification of a protein with homology to hsp90 that binds the type 1 tumor necrosis factor receptor. J. Biol. Chem. 270, 3574–3581.
Paull T.T., Rogakou E.P., Yamazaki V., Kirchgessner C.U., Gellert M., Bonner W.M. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10, 886–895.
Antequera F., Bird A. 1993. Number of CpG islands and genes in human and mouse. Proc. Natl. Acad. Sci. U. S. A. 90, 11995–11999.
Xu M., Cook P.R. 2008. Similar active genes cluster in specialized transcription factories. J. Cell Biol. 181, 615–623.
Gavrilov A.A., Zukher I.S., Philonenko E.S., Razin S.V., Iarovaia O.V. 2010. Mapping of the nuclear matrixbound chromatin hubs by a new M3C experimental procedure. Nucleic Acids Res. 38, 8051–8060.
Gavrilov A.A., Gushchanskaya E.S., Strelkova O., Zhironkina O., Kireev I.I., Iarovaia O.V., Razin S.V. 2013. Disclosure of a structural milieu for the proximity ligation reveals the elusive nature of an active chromatin hub. Nucleic Acids Res. 41, 3563–3575.
Razin S.V., Gavrilov A.A., Ioudinkova E.S., Iarovaia O.V. 2013. Communication of genome regulatory elements in a folded chromosome. FEBS Lett. 587, 1840–1847.
Nolis I.K., McKay D.J., Mantouvalou E., Lomvardas S., Merika M., Thanos D. 2009. Transcription factors mediate long-range enhancer-promoter interactions. Proc. Natl. Acad. Sci. U. S. A. 106, 20222–20227.
Marenduzzo D., Finan K., Cook P.R. 2006. The depletion attraction: An underappreciated force driving cellular organization. J. Cell Biol. 175, 681–686.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © E.S. Gushchanskaya, A.V. Artemov, S.V. Ulyanov, A.A. Penin, M.D. Logacheva, S.V. Razin, A.A. Gavrilov, 2014, published in Molekulyarnaya Biologiya, 2014, Vol. 48, No. 6, pp. 1008–1018.
Rights and permissions
About this article
Cite this article
Gushchanskaya, E.S., Artemov, A.V., Ulyanov, S.V. et al. Spatial organization of housekeeping genes in interphase nuclei. Mol Biol 48, 886–895 (2014). https://doi.org/10.1134/S0026893314060053
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893314060053