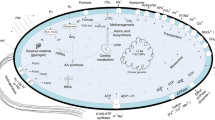
Abstract
Aerobic methanotrophic bacteria are prokaryotic microorganisms possessing methane monooxygenases, unique enzymes that determine their ability to utilize methane (CH4) as a growth substrate. This metabolic capability makes methanotrophs attractive objects for biotechnological applications aimed at utilizing methane for production of microbial cell protein and various target metabolites. The current raise of interest to these biotechnologies is driven by high availability of methane, which is a major component of natural gas, as well as of the biogas produced in anaerobic fermentation processes. Since aerobic methanotrophs oxidize methane at the ambient temperature and pressure, they represent natural cell factories for converting CH4 into various value-added products. Further development of biotechnologies based on methane utilization requires application of genome editing techniques to obtain producer strains with improved characteristics. For a long time, the progress in metabolic engineering of methanotrophs was hampered by their specific metabolic properties and the difficulties of handling these bacteria. Here, we present an overview of the latest achievements in the field of metabolic engineering of methanotrophic bacteria and identify the potential targets as well as the currently available tools for genome editing of these microorganisms. These techniques open up the possibility of constructing strains with biotechnologically relevant characteristics and conducting in-depth research of the metabolic features of aerobic methanotrophs.


Similar content being viewed by others
REFERENCES
Akberdin, I.R., Thompson, M., Hamilton, R., Desai, N., Alexander, D., Henard, C.A., Guarnieri, M.T., and Kalyuzhnaya, M.G., Methane utilization in Methylomicrobium alcaliphilum 20ZR: a systems approach, Sci. Rep., 2018, vol. 8, p. 2512.
Ali, H. and Murrell, J.C., Development and validation of promoter-probe vectors for the study of methane monooxygenase gene expression in Methylococcus capsulatus Bath, Microbiology (Reading), 2009, vol. 155, pp. 761‒771.
Bordel, S., Crombie, A.T., Muñoz, R., and Murrell, J.C., Genome Scale Metabolic Model of the versatile methanotroph Methylocella silvestris, Microb. Cell Fact., 2020, vol. 19, p. 144.
Borodina, E., Nichol, T., Dumont, M.G., Smith, T.J., and Murrell, J.C., Mutagenesis of the “leucine gate” to explore the basis of catalytic versatility in soluble methane monooxygenase, Appl. Environ. Microbiol., 2007, vol. 73, pp. 6460–6467.
Bosse, U. and Frenzel, P., Activity and distribution of methane-oxidizing bacteria in flooded rice soil microcosms and in rice plants (Oryza sativa), Appl. Environ. Microbiol., 1997, vol. 63, pp. 1199–1207.
Bothe, H., Moller Jensen, K., Mergel, A., Larsen, J., Jorgensen, C., Bothe, H., and Jorgensen, L., Heterotrophic bacteria growing in association with Methylococcus capsulatus (Bath) in a single cell protein production process, Appl. Microbiol. Biotechnol., 2002, vol. 59, pp. 33‒39.
But, S.Yu., Khmelenina, V.N., Mustakhimov, I.I., and Trotsenko, Yu.A., Construction and characterization of Methylomicrobium alcaliphilum 20Z knockout mutants defective in sucrose and ectoine biosynthesis genes, Microbiology (Moscow), 2013, vol. 82, pp. 253–255.
But, S.Y., Egorova, S.V., Khmelenina, V.N., and Trotsenko, Y.A. Biochemical properties and phylogeny of hydroxypyruvate reductases from methanotrophic bacteria with different C1-assimilation pathways, Biochemistry (Moscow), 2017, vol. 82, pp. 1295–1303.
But, S.Yu., Dedysh, S.N., Popov, V.O., Pimenov, N.V., and Khmelenina, V.N., Construction of a Type-I methanotroph with reduced capacity for glycogen and sucrose accumulation, Appl. Biochem. Microbiol., 2020a, vol. 56, pp. 538–543.
But, S.Y., Egorova, S.V., Khmelenina, V.N., and Mustakhimov, I.I., Malyl-CoA lyase provides glycine/glyoxylate synthesis in type I methanotrophs, FEMS Microbiol. Lett., 2020b, vol. 367, p. fnaa207.
Chaumeil, P.-A., Mussig, A.J., Hugenholtz, P., and Parks, D.H., GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database, Bioinformatics, 2020, vol. 36, pp. 1925–1927.
Chistoserdova, L. and Lidstrom, M.E., Aerobic methylotrophic prokaryotes, in The Prokaryotes: Prokaryotic Physiology and Biochemistry, Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., and Thompson, F., Eds., Berlin Heidelberg: Springer, 2013, pp. 267–285.
Chu, F., Beck, D.C., and Lidstrom, M.E., MxaY regulates the lanthanide-mediated methanol dehydrogenase switch in Methylomicrobium buryatense, PeerJ., 2016, vol. 4, p. e2435.
Chu, F. and Lidstrom, M.E., XoxF acts as the predominant methanol dehydrogenase in the type I methanotroph Methylomicrobium buryatense, J. Bacteriol., 2016, vol. 198, pp. 1317–1325.
Conrad, R., The global methane cycle: recent advances in understanding the microbial processes involved, Environ. Microbiol. Rep., 2009, vol. 1, pp. 285–292.
Crombie, A. and Murrell, J.C., Development of a system for genetic manipulation of the facultative methanotroph Methylocella silvestris BL2, Methods Enzymol., 2011, vol. 495, pp. 119‒133.
Csaki, R., Bodrossy, L., Klem, J., Murrell, J.C., and Kovacs, K.L., Genes involved in the copper-dependent regulation of soluble methane monooxygenase of Methylococcus capsulatus (Bath): cloning, sequencing and mutational analysis, Microbiology (Reading), 2003, vol. 149, pp. 1785–1795.
Davamani, V., Parameswari, E., and Arulmani, S., Mitigation of methane gas emissions in flooded paddy soil through the utilization of methanotrophs, Sci. Total Environ., 2020, vol. 726, p. 138570.
De la Torre, A., Metivier, A., Chu, F., Laurens, L.M., Beck, D.A., Pienkos, P.T., Lidsrom, M.E., and Kalyuzhnaya, M.G., Genome-scale metabolic reconstructions and theoretical investigation of methane conversion in Methylomicrobium buryatense strain 5G(B1), Microb. Cell Fact., 2015, vol. 14, p. 188.
Dedysh, S.N. and Dunfield, P.F., Facultative methane oxidizers, in Handbook of Hydrocarbon and Lipid Microbiology, Timmis, K.N., Ed., Berlin: Springer-Verlag, 2010, pp. 1967‒1976.
Dedysh, S.N. and Knief, C., Diversity and phylogeny of described aerobic methanotrophs, in Methane Biocatalysis: Paving the Way to Sustainability, Kalyuzhnaya, M. and Xing, X.H., Eds., Cham: Springer, 2018, pp. 17–42.
Dunfield, P.F. and Dedysh, S.N., Methylocella: a gourmand among methanotrophs, Trends Microbiol., 2014, vol. 22, pp. 368‒369.
Eccleston, M. and Kelly, D.P., Assimilation and toxicity of some exogenous C1 compounds, alcohols, sugars and acetate in the methane-oxidizing bacterium Methylococcus capsulatus, J. Gen. Microbiol., 1973, vol. 75, pp. 211‒221.
Egorov, I., Kupina, L., Aksyuk, I., and Murtazaeva, R., Gaprin—a protein source, Ptitsevidstvo, 1990, vol. 8, pp. 25‒27.
Eshinimaev, B.T., Khmelenina, V.N., Sakharovskii, V.G., Suzina, N.E., and Trotsenko, Y.A., Physiological, biochemical, and cytological characteristics of a haloalkalitolerant methanotroph grown on methanol, Microbiology (Moscow), 2002, vol. 71, pp. 512‒518.
Espah Borujeni, A., Channarasappa, A.S., and Salis, H.M., Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites, Nucleic Acids Res., 2014, vol. 42, pp. 2646‒2659.
Fu, Y., He, L., Reeve, J., Beck, D.A.C., and Lidstrom, M.E., Core metabolism shifts during growth on methanol versus methane in the methanotroph Methylomicrobium buryatense 5GB1, MBio, 2019, vol. 10, p. e00406-19.
Fu, Y., Li, Y., and Lidstrom, M., The oxidative TCA cycle operates during methanotrophic growth of the Type I methanotroph Methylomicrobium buryatense 5GB1, Metab. Eng., 2017, vol. 42, pp. 43–51.
Gal’chenko, V.F., Metanotrofnye bakterii (Methanotrophic Bacteria), Moscow: GEOS.
Garg, S., Wu, H., Clomburg, J.M., and Bennett, G.N., Bioconversion of methane to C-4 carboxylic acids using carbon flux through acetyl-CoA in engineered Methylomicrobium buryatense 5GB1C, Metab. Eng., 2018, vol. 48, pp. 175‒183.
Garneau, J.E., Dupuis, M.-È., Villion, M., Romero, D.A., Barrangou, R., Boyaval, P., Fremaux, C., Horvath, P., Magadán, A.H., and Moineau, S., The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA, Nature, 2010, vol. 468, pp. 67‒71.
Gasiunas, G., Barrangou, R., Horvath, P., and Siksnys, V., Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria, Proc. Natl. Acad. Sci. USA, 2012, vol. 109, pp. E2579–E2586.
Grigoryan, A.N. and Gorskaya, L.A., Ispol’zovanie prirodnogo gaza dlya mikrobiologicheskogo sinteza (Utilization of Natural Gas for Microbiological Synthesis), Moscow, ONTI Mikrobiopriom, 1970.
Haque, M.F.U., Gu, W., DiSpirito, A.A., and Semrau, J.D., Marker exchange mutagenesis of mxaF, encoding the large subunit of the Mxa methanol dehydrogenase, in Methylosinus trichosporium OB3b, Appl. Environ. Microbiol., 2016, vol. 82, pp. 1549–1555.
Hamer, G. and Harrison, D.E.F., Single cell protein: the technology, economics and future potential, in Hydrocarbons in Biotechnology, Harrison, D.E.F., Higgins, I.J., and London, W.R., Eds., London: Heyden Institute of Petroleum, 1980, pp. 59–73.
Hanson, R.S. and Hanson, T.E., Methanotrophic bacteria, Microbiol. Rev., 1996, vol. 60, pp. 439‒471.
Harwood, J.H., Williams, E., and Bainbridge, B.W., Mutation of the methane oxidizing bacterium Methylococcus capsulatus, J. Appl. Bacteriol., 1972, vol. 35, pp. 99–108.
Henard, C.A., Franklin, T.G., Youhenna, B., But, S., Alexander, D., Kalyuzhnaya, M.G., and Guarnieri, M.T., Biogas biocatalysis: methanotrophic bacterial cultivation, metabolite profiling, and bioconversion to lactic acid, Front. Microbiol., 2018, vol. 9, p. 2610.
Henard, C.A., Smith, H., Dowe, N., Kalyuzhnaya, M.G., Pienkos, P.T., and Guarnieri, M.T., Bioconversion of methane to lactate by an obligate methanotrophic bacterium, Sci. Rep., 2016, vol. 6, p. 21585.
Henard, C.A., Wu, C., Xiong, W., Henard, J.M., Davidheiser-Kroll, B., Orata, F.D., and Guarnieri, M.T., Ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) is essential for growth of themethanotroph Methylococcus capsulatus strain Bath, Appl. Environ. Microbiol., 2021, vol. 87, p. e00881–21.
Henard, C.A., Bourret, T.J., Song, M., and Vázquez-Torres, A., Control of redox balance by the stringent response regulatory protein promotes antioxidant defenses of Salmonella, J. Biol. Chem., 2010, vol. 285, pp. 36785–36793.
Henard, C.A., Akberdin, I.R., Kalyuzhnaya, M.G., and Guarnieri, M.T., Muconic acid production from methane using rationally-engineered methanotrophic biocatalysts, Green Chem., 2019, vol. 21, pp. 6731–6737.
Hu, L., Guo, S., Yan, X., Zhang, T., Xiang, J., and Fei, Q., Exploration of an efficient electroporation system for heterologous gene expression in the genome of methanotroph, Front. Microbiol., 2021, vol. 12, p. 717033.
Ishikawa, M., Yokoe, S., Kato, S., and Hori, K., Efficient counterselection for Methylococcus capsulatus (Bath) by using a mutated pheS gene, Appl. Environ. Microbiol., 2018, vol. 84, p. e01875-18.
Jiang, D., Kim, C.S., Hanson, R.S., and Wood, T.K., Optimization of trichloroethylene degradation using soluble methane monooxygenase of Methylosinus trichosporium OB3b expressed in recombinant bacteria, Biotechnol. Bioeng., 1996, vol. 51, pp. 349–359.
Jiang, H., Chen, Y., Jiang, P., Zhang, C., Smith, T.J., Murrell, J.C., and Xing, X.H., Methanotrophs: multifunctional bacteria with promising applications in environmental bioengineering, Biochem. Eng J., 2010, vol. 49, pp. 277‒288.
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J.A., and Charpentier, E., A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity, Science, 2012, vol. 337, pp. 816–821.
Kalyuzhnaya, M.G., Kumaresan, D., Heimann, K., Caetano, N.S., Visvanathan, C., and Parthiba Karthikeyan, O., Editorial: Methane: a bioresource for fuel and biomolecules, Front. Environ. Sci., 2020, vol. 8, p. 9.
Kalyuzhnaya, M.G., Puri, A.W., and Lidstrom, M.E., Metabolic engineering in methanotrophic bacteria, Metab. Engin., 2015, vol. 29, pp. 142‒152.
Kalyuzhnaya, M.G., Yang, S., Rozova, O.N., Smalley, N.E., Clubb, J., Lamb, A., Gowda, G.N., Raftery, D., Fu, Y., Bringel, F., Vuilleumier, S., Beck, D.A.C., Trotsenko, Y.A., Khmelenina, V.N., and Lidstrom, M.E., Highly efficient methane biocatalysis revealed in a methanotrophic bacterium, Nat. Commun., 2013, vol. 4, p. 2785.
Khadem, A.F., Pol, A., Wieczorek, A., Mohammadi, S.S., Francoijs, K.J., Stunnenberg, H.G., Jetten, M.S.M., and Op den Camp, H.J.M., Autotrophic methanotrophy in verrucomicrobia: Methylacidiphilum fumariolicum SolV uses the Calvin‒Benson‒Bassham cycle for carbon dioxide fixation, J. Bacteriol., 2011, vol. 193, pp. 4438–4446.
Khider, M.L.K., Brautaset, T., and Irla, M., Methane monooxygenases: central enzymes in methanotrophy with promising biotechnological applications, World J. Microbiol. Biotechnol., 2021, vol. 37, p. 72.
Khmelenina, V.N., But, S.Y., Rozova, O.N., and Trotsenko, Y.A., Metabolic features of aerobic methanotrophs: news and views, Curr. Issues Mol. Biol., 2019, vol. 33, pp. 85‒100.
Khmelenina, V.N., Kalyuzhnaya, M.G., Sakharovsky, V.G., Suzina, N.E., Trotsenko, Y.A., and Gottschalk, G., Osmoadaptation in halophilic and alkaliphilic methanotrophs, Arch. Microbiol., 1999, vol. 172, pp. 321‒329.
Knief, C., Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker, Front. Microbiol., 2015, vol. 6, p. 1346.
Lalov, V.V., Analysis and synthesis of energotechnological systems of fodder protein production from natural gas, Extended Abstract of Doctoral (Biology) Dissertation, Moscow, 1991.
Le, H.T.Q., Nguyen, A.D., Park, Y.R., and Lee, E.Y., Sustainable biosynthesis of chemicals from methane and glycerol via reconstruction of multi-carbon utilizing pathway in obligate methanotrophic bacteria, Microb. Biotechnol., 2021, vol. 14, pp. 2552‒2565.
Lee, J.K., Kim, S., Kim, W., Kim, S., Cha, S., Moon, H., Hur, D.H., Kim, S.Y., Na, J.G., Lee, J.W., Lee, E.Y., and Hahn, J.S., Efficient production of d-lactate from methane in a lactate-tolerant strain of Methylomonas sp. DH-1 generated by adaptive laboratory evolution, Biotechnol. Biofuels, 2019, vol. 12, pp. 234.
Lee, H.M., Ren, J., Tran, K.M., Jeon, B.M., Park, W.U., Kim, H., Lee, K.E., Oh, Y., Choi, M., Kim, D.S., and Na, D., Identification of efficient prokaryotic cell-penetrating peptides with applications in bacterial biotechnology, Commun. Biol., 2021a, vol. 4, p. 205.
Lee, H.M., Ren, J., Yu, M.S., Kim, H., Kim, W.Y., Shen, J., Yoo, S.M., Eyun, S.I., and Na, D., Construction of a tunable promoter library to optimize gene expression in Methylomonas sp. DH-1, a methanotroph, and its application to cadaverine production, Biotechnol. Biofuels, 2021b, vol. 14, p. 228.
Lieven, C., Petersen, L.A.H., Jørgensen, S.B., Gernaey, K.V., Herrgard, M.J., and Sonnenschein, N., A genome-scale metabolic model for Methylococcus capsulatus (Bath) suggests reduced efficiency electron transfer to the particulate methane monooxygenase, Front. Microbiol., 2018, vol. 9, p. 2947.
Liu, Y., He, X., Zhu, P., Cheng, M., Hong, Q., and Yan, X., pheSAG based rapid and efficient markerless mutagenesis in Methylotuvimicrobium, Front. Microbiol., 2020, vol. 11, p. 441.
Liu, Y., Zhang, H., He, X., and Liu, J., Genetically engineered methanotroph as a platform for bioaugmentation of chemical pesticide contaminated soil, ACS Synth. Biol., 2021, vol. 10, pp. 487–494.
Lloyd, J.S., Finch, R., Dalton, H., and Murrell, J.C. Homologous expression of soluble methane monooxygenase genes in Methylosinus trichosporium OB3b, Microbiology (Reading), 1999, vol. 145, pp. 461‒470.
Marx, C.J. and Lidstrom, M.E., Development of improved versatile broad-host-range vectors for use in methylotrophs and other gram-negative bacteria, Microbiology, 2001, vol. 147, pp. 2065‒2075.
Marx, C.J. and Lidstrom, M.E., Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria, Biotechniques, 2002, vol. 33, pp. 1062‒1067.
Marx, C.J., Development of a broad-host-range sacB-based vector for unmarked allelic exchange, BMC Res. Notes, 2008, vol. 1, p. 1. https://doi.org/10.1186/1756-0500-1-1
Murrell, J.C., Gilbert, B., and McDonald, I.R., Molecular biology and regulation of methane monooxygenase, Arch. Microbiol., 2000, vol. 173, pp. 325–332.
Murrell, J.C. and Smith, T.J., Biochemistry and molecular biology of methane monooxygenase, in Handbook of Hydrocarbon and Lipid Microbiology, Timmis, K.N., Ed., Berlin‒Heidelberg: Springer, 2010, pp. 1045–1055.
Mustakhimov, I.I., Reshetnikov, A.S., Glukhov, A.S., Khmelenina, V.N., Kalyuzhnaya, M.G., and Tro-tsenko, Y.A., Identification and characterization of EctR1, a new transcriptional regulator of the ectoine biosynthesis genes in the halotolerant methanotroph Methylomicrobium alcaliphilum 20Z, J. Bacteriol., 2010, vol. 192, pp. 410–417.
Mustakhimov, I.I., But, S.Y., Reshetnikov, A.S., Khmelenina, V.N., and Trotsenko, Y.A., Homo- and heterologous reporter proteins for evaluation of promoter activity in Methylomicrobium alcaliphilum 20Z, Appl. Biochem. Microbiol., 2016, vol. 52, pp. 279‒286.
Naizabekov, S. and Lee, E.Y., Genome-scale metabolic model reconstruction and in silico investigations of methane metabolism in Methylosinus trichosporium OB3b, Microorganisms, 2020, vol. 8, p. 437.
Nguyen, A.D., Hwang, I.Y., Lee, O.K., Kim, D., Kalyuzhnaya, M.G., Mariyana, R., Hadiyati, S., Kim, M.S., and Lee, E.Y., Systematic metabolic engineering of Methylomicrobium alcaliphilum 20Z for 2,3-butanediol production from methane, Metab. Engin., 2018, vol. 47, pp. 323‒333.
Nguyen, L.T. and Lee, E.Y., Biological conversion of methane to putrescine using genome-scale model-guided metabolic engineering of a methanotrophic bacterium Methylomicrobium alcaliphilum 20Z, Biotechnol. Biofuels, 2019, vol. 12, p. 147.
Nguyen, D.T.N., Lee, O.K., Hadiyati, S., Affifah, A.N., Kim, M.S., and Lee, E.Y., Metabolic engineering of the type I methanotroph Methylomonas sp. DH-1 for production of succinate from methane, Metab. Engin., 2019, vol. 54, pp. 170‒179.
Nguyen, D., Lee, O.K., Lim, C., Lee, J., Na, J.-G., and Lee, E.Y., Metabolic engineering of type II methanotroph, Methylosinus trichosporium OB3b, for production of 3-hydroxypropionic acid from methane via a malonyl-CoA reductase-dependent pathway, Metab. Engin., 2020a, vol. 59, pp. 142‒150.
Nguyen, A.D., Kim, D., and Lee, E.Y., Unlocking the biosynthesis of sesquiterpenoids from methane via the methylerythritol phosphate pathway in methanotrophic bacteria, using α-humulene as a model compound, Metab. Engin., 2020b, vol. 61, pp. 69–78.
Nguyen, A.D., Chau, T.H.T., and Lee, E.Y., Methanotrophic microbial cell factory platform for simultaneous conversion of methane and xylose to value-added chemicals, Chem. Eng. J., 2021, vol. 420, p. 127632.
Op den Camp, H.J.M., Islam, T., Stott, M.B., Harhangi, H.R., Hynes, A., Schouten, S., Jetten, M.S.M., Birkeland, N.K., Pol, A., and Dunfield, P.F., Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia, Environ. Microbiol. Rep., 2009, vol. 1, pp. 293–306.
Øverland, M., Tauson, A.H., Shearer, K., and Skrede, A., Evaluation of methane-utilising bacteria products as feed ingredients for monogastric animals, Arch. Anim. Nutr., 2010, vol. 64, pp. 171‒189.
Parks, D.H., Chuvochina, M., Rinke, C., Mussig, A.J., Chaumeil, P.-A., and Hugenholtz, P., GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy, Nucleic Acids Res., 2022, vol. 50, pp. D785–D794.
Pham, D.N., Nguyen, A.D., and Lee, E.Y., Outlook on engineering methylotrophs for one-carbon-based industrial biotechnology, Chem. Eng. J., 2022a, vol. 449, p. 137769.
Pham, D.N., Nguyen, A.D., Oh, S.H., and Lee, E.Y., Bypassing the bottlenecks in the shikimate and methylerythritol phosphate pathways for enhancing the production of natural products from methane in Methylotuvimicrobium alcaliphilum 20Z, Green Chem., 2022b, vol. 24, pp. 2893‒2903.
Pham, D.N., Mai, D.H.A., Nguyen, A.D., Chau, T.H.T., and Lee, E.Y., Development of an engineered methanotroph-based microbial platform for biocatalytic conversion of methane to phytohormone for sustainable agriculture, Chem. Eng. J., 2022c, vol. 429, p. 132522.
Pieja, A.J., Morse, M.C., and Cal, A.J., Methane to bioproducts: the future of the bioeconomy?, Curr. Opin. Chem. Biol., 2017, vol. 41, pp. 123–131.
Puri, A.W., Owen, S., Chu, F., Chavkin, T., Beck, D.A.C., and Kalyuzhnaya, M.G., Genetic tools for the industrially promising methanotroph Methylomicrobium buryatense, Appl. Environ. Microbiol., 2015, vol. 81, pp. 1775–1781.
Recorbet, G., Robert, C., Givaudan, A., Kudla, B., Normand, P., and Faurie, G., Conditional suicide system of Escherichia coli released into soil that uses the Bacillus subtilis sacB gene, Appl. Environ. Microbiol., 1993, vol. 59, pp. 1356‒1365.
Ren, J., Lee, H.‑M., Thai, T.D., and Na, D., Identification of a cytosine methyltransferase that improves transformation efficiency in Methylomonas sp. DH‑1, Biotechnol. Biofuels, 2020, vol. 13, p. 200.
Ro, S.Y. and Rosenzweig, A.C., Recent advances in the genetic manipulation of Methylosinus trichosporium OB3b, Methods Enzymol., 2018, vol. 605, pp. 335‒349.
Salis, H.M., Mirsky, E.A. and Voigt, C.A., Automated design of synthetic ribosome binding sites to control protein expression, Nat. Biotechnol., 2009, vol. 27, pp. 946‒950.
Schmitz, R.A., Peeters, S.H., Versantvoort, W., Picone, N., Pol, A., Jetten, M.S.M., and Op Den Camp, H.J.M., Verrucomicrobial methanotrophs: ecophysiology of metabolically versatile acidophiles, FEMS Microbiol. Rev., 2021, vol. 45, p. fuab007.
Shaner, N.C., Campbell, R.E., Steinbach, P.A., Giepmans, B.N.G., Palmer, A.E., and Tsien, R.Y., Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein, Nat. Biotechnol., 2004, vol. 22, pp. 1567‒1572.
Stafford, G.P., Scanlan, J., McDonald, I.R., and Murrell, J.C., rpoN, mmoR and mmoG, genes involved in regulating the expression of soluble methane monooxygenase in Methylosinus trichosporium OB3b, Microbiology (Reading), 2003, vol. 149, pp. 1771–1784.
Sternberg, N. and Hamilton, D., Bacteriophage P1 site-specific recombination: I. Recombination between loxP sites, J. Mol. Biol., 1981, vol. 150, pp. 467‒486.
Strong, P.J., Xie, S., and Clarke, W.P., Methane as a resource: can the methanotrophs add value?, Environ Sci. Technol., 2015, vol. 49, pp. 4001–4018.
Tapscott, T., Guarnieri, M.T., and Henard, C.A., Development of a CRISPR/Cas9 system for Methylococcus capsulatus in vivo gene editing, Appl. Environ. Microbiol., 2019, vol. 85, p. e00340-19.
Trotsenko, Y.A. and Murrell, J.C., Metabolic aspects of aerobic obligate methanotrophy, Adv. Appl. Microbiol., 2008, vol. 63, pp. 183–229.
Welander, P.V. and Summons, R.E., Discovery, taxonomic distribution, and phenotypic characterization of a gene required for 3-methylhopanoid production, Proc. Natl. Acad. Sci. USA, 2012, vol. 109, pp. 12905–12910.
Williams, E., Shimmin, M.S., and Bainbridge, B.W., Mutation in the obligate methylotrophs Methylococcus capsulatus and Methylomonas albus, FEMS Microbiol. Lett., 1977, vol. 2, pp. 293–296.
Wilson, E.H., Groom, J.D., Sarfatis, M.C., Ford, S.M., Lidstrom, M.E., and Beck, D.A., A computational framework for identifying promoter sequences in nonmodel organisms using RNA-seq data sets, ACS Synth. Biol., 2021, vol. 10, pp. 1394–1405.
Yan, X., Chu, F., Puri, A.W., Fu, Y., and Lidstrom, M.E., Electroporation-based genetic manipulation in type I methanotrophs, Appl. Environ. Microbiol., 2016, vol. 82, pp. 2062–2069.
Ye, R.W., Yao, H., Stead, K., Wang, T., Tao, L., Cheng, Q., Sharpe, P.L., Suh, W., Nagel, E., Arcilla, D., Dragotta, D., and Miller, E.S., Construction of the astaxanthin biosynthetic pathway in a methanotrophic bacterium Methylomonas sp. strain 16a, Ind. Microbiol. Biotechnol., 2007, vol. 34, pp. 289‒299.
Zheng, Y., Huang, J., Zhao, F., and Chistoserdova, L., Physiological effect of XoxG(4) on lanthanide-dependent methanotrophy, mBio, 2018, vol. 9, p. e02430-17.
Funding
The review was prepared as a part of the project no. 075-15-2021-1071 “Development of Genomic Editing Techniques for the Purposes of Innovation in Industrial and Food Biotechnologies” with financial support of the Ministry of Science and Higher Education of the Russian Federation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest. The authors declare that they have no conflict of interests.
Statement on the welfare of animals. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by D. Timchenko
Rights and permissions
About this article
Cite this article
Khmelenina, V.N., But, S.Y., Rozova, O.N. et al. Genome Editing in Methanotrophic Bacteria: Potential Targets and Available Tools. Microbiology 91, 613–630 (2022). https://doi.org/10.1134/S0026261722602196
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026261722602196