Abstract—
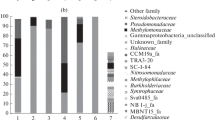
The technology of single-cell protein production from natural gas is based on using thermotolerant methanotrophic bacteria with high growth rates on methane. So far, the spectrum of strains used for industrial purposes was restricted to members of the genus Methylococcus. This poses limitations to further development of this technology and fuels the search for new cultures of fast-growing methanotrophs. The later task was addressed in the present work by analyzing the sediment samples of the Chernaya River, Crimea. Molecular analysis of the microbial community composition in the sediment revealed Gammaproteobacteria as the predominant group (33‒42% of all retrieved 16S rRNA gene fragments), as well as Chloroflexi, Actinobacteria, Alphaproteobacteria, Bacteroidota, and Acidobacteria as other numerically significant community members. The methanotrophic enrichment culture obtained from the sediment contained bacteria of the genus Methylomonas as the major component, with the relative abundance of up to 60% of all 16S rRNA gene fragments. The use of various strategies for methanotroph isolation resulted in obtaining three isolates of target bacteria of the genera Methylomonas, Methylomagnum and Methylocystis. The optimal growth temperatures of these isolates were 25, 35, and 40°С, respectively. The highest specific growth rate in batch culture, 0.21 h−1, was determined for Methylocystis sp. Kr9, which displayed 99.22 and 99.13% 16S rRNA gene sequence similarity to the type strains of two Methylocystis species, Methylocystis echinoides IMET 10491T and Methylocystis parvus OBBPT, respectively.



Similar content being viewed by others
REFERENCES
Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J., Gapped BLAST and PSI-BLAST: a new generation of protein, Nucleic Acids Res., 1997, vol. 25, pp. 3389–3402.
Auman, A.J., Stolyar, S., Costello, A.M., and Lidstrom, M.E., Molecular characterization of methanotrophic isolates from freshwater lake sediment, Appl. Environ. Micro-biol., 2000, vol. 66, pp. 5259–5266.
Beck, D.A.C., Kalyuzhnaya, M.G., Malfatti, S., Tringe, S.G., del Rio, T.G., Ivanova, N., Lidstrom, M.E., and Chistoserdova, L., A metagenomic insight into freshwater methane-utilizing communities and evidence for cooperation between the Methylococcaceae and the Methylophilaceae, PeerJ., 2013, vol. 1:e23.
Bol’shakov, A.M. and Egorov, A.V., Application of the phase equilibrium degassing in gasometric research, Okeanologiya, 1987, vol. 27, no. 5, pp. 861–862.
Bolyen, E., Rideout, J.R., Dillon, M.R., Bokulich, N.A., Abnet, C.C., Al-Ghalith, G.A., Alexander, H., Alm, E.J., Arumugam, M., Asnicar, F., Bai, Y., Bisanz, J.E., Bittinger, K., Brejnrod, A., Brislawn, C.J., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2, Nat. Biotechnol., 2019, vol. 37, pp. 852–857.
Callahan, B.J., McMurdie, P.J., Rosen, M.J., Han, A.W., Johnson, A.J.A., and Holmes, S.P., DADA2: high-resolution sample inference from Illumina amplicon data, Nat. Methods, 2016, vol. 13, pp. 581–583.
Cébron, A., Bodrossy, L., Chen, Y., Singer, A.C., Thompson, I.P., Prosser, J.I., and Murrell, J.C., Identity of active methanotrophs in landfill cover soil as revealed by DNA-stable isotope probing, FEMS Microbiol. Ecol., 2007, vol. 62, pp. 12–23.
Conrado, R.J. and Gonzalez, R., Envisioning the bioconversion of methane to liquid fuels, Science, 2014, vol. 343, pp. 621–623.
Conrad, R., The global methane cycle: recent advances in understanding the microbial processes involved, Environ. Microbiol. Rep., 2009, vol. 1, pp. 285–292.
Costello, A.M., Auman, A.J., Macalady, J.L., Scow, K.M., and Lidstrom, M.E., Estimation of methanotroph abundance in a freshwater lake sediment, Environ. Microbiol., 2002, vol. 4, pp. 443–450.
Coty, V., A critical review of the utilization of methane, Biotech. Bioeng. Symp., 1969, vol. 1, pp. 105–117.
Danilova, O.V., Belova, S.E., Gagarinova, I.V., and Dedysh, S.N., Microbial community composition and methanotroph diversity of a subarctic wetland in Russia, Microbiology (Moscow), 2016, vol. 85, pp. 583–591.
Danilova, O.V., Kulichevskaya, I.S., Rozova, O.N., Detkova, E.N., Bodelier, P.L.E., Trotsenko, Y.A., and Dedysh, S.N., Methylomonas paludis sp. nov., the first acidtolerant member of the genus Methylomonas, from an acidic wetland, Int. J. Syst. Evol. Microbiol., 2013, vol. 63, no. 6, pp. 2282–2289.
Dedysh, S.N. and Knief, C., Diversity and phylogeny of described aerobic methanotrophs, in Methane Biocatalysis: Paving the Way to Sustainability, Kalyuzhnaya, M. and Xing, X., Eds., Springer, Cham, 2018, pp. 17–42.
Dedysh, S.N., Exploring methanotroph diversity in acidic northern wetlands: molecular and cultivation-based studies, Microbiology (Moscow), 2009, vol. 78, pp. 655–669.
Deutzmann, J.S., Hoppert, M., and Schink, B., Characterization and phylogeny of a novel methanotroph, Methyloglobulus morosus gen. nov., spec. nov, Syst. Appl. Microbiol., 2014, vol. 37, pp. 165–169.
Deutzmann, J.S., Wörner, S., and Schink, B., Activity and diversity of methanotrophic bacteria at methane seeps in eastern Lake Constance sediments, Appl. Environ. Microbiol., 2011, vol. 77, pp. 2573–2581.
Gal’chenko, V.F., Metanotrofnye bakterii (Methanotrophic Bacteria), Moscow: GEOS, 2001.
Ganendra, G., Wang, J., Ramos, J.A., Derluyn, H., Rahier, H., Cnudde, V., Ho, A., and Boon, N., Biogenic concrete protection driven by the formate oxidation by Methylocystis parvus OBBP, Front. Microbiol., 2015, vol. 6, p. 786.
Grigoryan, A.N. and Gorskaya, L., Ispol’zovanie prirodnogo gaza dlya mikrobiologicheskogo sinteza (Application of Natural Gas for Microbiological Synthesis), Moscow: ONTI Mikrobioprom, 1970.
Guo, W., Li, D., He, R., Wu, M., Chen, W., Gao, F., Zhang, Z., Yao, Y., Yu, L., and Chen, S., Synthesizing value-added products from methane by a new Methylomonas, J. Appl. Microbiol., 2017, vol. 123, pp. 1214–1227.
Hanson, R. and Hanson, T., Methanotrophic bacteria, Microbiol. Rev., 1996, vol. 60, pp. 439–471.
Hirayama, H., Abe, M., Miyazaki, M., Nunoura, T., Furushima, Y., Yamamoto, H., and Takai, K., Methylomarinovum caldicuralii gen. nov., sp. nov., a moderately thermophilic methanotroph isolated from a shallow submarine hydrothermal system, and proposal of the family Methylothermaceae fam. nov., Int. J. Syst. Evol. Microbiol., 2014, vol. 64, pp. 989‒999.
Hoefman, S., van der Ha, D., De Vos, P., Boon, N., and Heylen, K., Miniaturized extinction culturing is the preferred strategy for rapid isolation of fast-growing methane-oxidizing bacteria, Microb. Biotechnol., 2012, vol. 5, pp. 368–378.
Houghton, K.M. and Stewart, L.C., Temperature-gradient incubation isolates multiple competitive species from a single environmental sample, Access Microbiol., 2020, vol. 2, pp. 1–9.
Joergensen, L. and Degn, H., Growth rate and methane affinity of a turbidostatic and oxystatic continuous culture of Methylococcus capsulatus (Bath), Biotechnol. Lett., 1987, vol. 9, pp. 71–76.
Kalyuzhnaya, M.G., Kumaresan, D., Heimann, K., Caetano, N.S., Visvanathan, C., and Parthiba Karthikeyan, O., Editorial: methane: a bioresource for fuel and biomolecules, Front. Environ. Sci., 2020, vol. 8, pp. 8–10.
Khalifa, A., Gyu Lee, C., Ogiso, T., Ueno, C., Dianou, D., Demachi, T., Katayama, A., and Asakawa, S., Methylomagnum ishizawai gen. nov., sp. nov., a mesophilic type I methanotroph isolated from rice rhizosphere, Int. J. Syst. Evol. Microbiol., 2015, vol. 65, pp. 3527–3534.
Khmelenina, V.N., Murrell, J.C., Smith, T.J., and Trotsenko, Y.A., Physiology and biochemistry of the aerobic methanotrophs, in Aerobic Utilization of Hydrocarbons, Oils and Lipids. Handbook of Hydrocarbon and Lipid Microbiology, Rojo, F., Ed., Springer, Cham., 2018, pp. 1–25.
Kim, J., Kim, D.H.D., and Yoon, S., Rapid isolation of fast-growing methanotrophs from environmental samples using continuous cultivation with gradually increased dilution rates, Appl. Microbiol. Biotechnol., 2018, vol. 102, pp. 5707–5715.
Knief, C., Diversity and habitat preferences of cultivated and uncultivated methanotrophic bacteria evaluated based on pmoA as molecular marker, Front. Microbiol., 2015, vol. 6, art. 1346.
Kumaresan, D., Abell, G.C.J., Bodrossy, L., Stralis-Pavese, N., and Murrell, J.C., Spatial and temporal diversity of methanotrophs in a landfill cover soil are differentially related to soil abiotic factors, Environ. Microbiol. Rep., 2009, vol. 1, pp. 398–407.
Ma, K., Conrad, R., and Lu, Y., Dry/wet cycles change the activity and population dynamics of methanotrophs in rice field soil, Appl. Environ. Microbiol., 2013, vol. 79, pp. 4932–4939.
Morinaga, Y., Yamanaka, S., Otsuka, S.I., and Hirose, Y., Characteristics of a newly isolated methane oxidizing bacterium, Methylomonas flagellata nov. sp., Agric. Biol. Chem., 1976, vol. 40, pp. 1539–1545.
Murrell, J.C., The aerobic methane oxidizing bacteria (Methanotrophs) BT, in Handbook of Hydrocarbon and Lipid Microbiology, Timmis, K.N., Ed., Heidelberg: Springer, 2010, pp. 1953–1966.
Pieja, A.J., Sundstrom, E.R., and Criddle, C.S., Poly-3-hydroxybutyrate metabolism in the type II methanotroph Methylocystis parvus OBBP, Appl. Environ. Microbiol., 2011, vol. 77, pp. 6012–6019.
Plyasov, Yu.M., Integrated assessment of nutritional value of microbial feed protein, Biotekhnologiya, 1988, vol. 4, no. 3, pp. 402–408.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., Peplies, J., and Glöckner, F.O., The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools, Nucleic Acids Res., 2013, vol. 41, pp. 590–596.
Rahalkar, M., Deutzmann, J., Schink, B., and Bussmann, I., Abundance and activity of methanotrophic bacteria in littoral and profundal sediments of Lake Constance (Germany), Appl. Environ. Microbiol., 2009, vol. 75, pp. 119–126.
Ritala, A., Häkkinen, S.T., Toivari, M., and Wiebe, M.G., Single cell protein-state-of-the-art, industrial landscape and patents 2001‒2016, Front. Microbiol., 2017, vol. 8, p. 2009.
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F., VSEARCH: a versatile open source tool for metagenomics, PeerJ., 2016, vol. 4, e2584.
Rostkowski, K.H., Pfluger, A.R., and Criddle, C.S., Stoichiometry and kinetics of the PHB-producing type II methanotrophs Methylosinus trichosporium OB3b and Methylocystis parvus OBBP, Bioresour. Technol., 2013, vol. 132, pp. 71–77.
Rumah, B.L., Stead, C.E., Claxton Stevens, B.H., Minton, N.P., Grosse-Honebrink, A., and Zhang, Y., Isolation and characterisation of Methylocystis spp. for poly-3-hydroxybutyrate production using waste methane feedstocks, AMB Express, 2021, vol. 11, art. 6.
Strong, P.J., Kalyuzhnaya, M., Silverman, J., and Clarke, W.P., A methanotroph-based biorefinery: Potential scenarios for generating multiple products from a single fermentation, Bioresour. Technol., 2016, vol. 215, pp. 314–323.
Strong, P.J., Xie, S., and Clarke, W.P., Methane as a resource: can the methanotrophs add value?, Environ. Sci. Technol., 2015, vol. 49, pp. 4001–4018.
Sundstrom, E.R. and Criddle, C.S., Optimization of methanotrophic growth and production of poly(3-hydroxybutyrate) in a high-throughput microbioreactor system, Appl. Environ. Microbiol., 2015, vol. 81, pp. 4767–4773.
Trotsenko, Y.A. and Murrell, J.C., Metabolic aspects of aerobic obligate methanotrophy, Adv. Appl. Microbiol., 2008, vol. 63, pp. 183–229.
Trotsenko, Y.A., Medvedkova, K.A., Khmelenina, V.N., and Eshinimayev, B.Ts., Thermophilic and thermotolerant aerobic methanotrophs, Microbiology (Moscow), 2009, vol. 78, pp. 387–401.
Trotsenko, Yu.A. and Khmelenina, V.N., Ekstremofil’nye metanotrofy (Extremophilic Methanotrophs), Moscow: ONTI PNTs RAN, 2009.
Vaksmaa, A., van Alen, T.A., Ettwig, K.F., Lupotto, E., Valè, G., Jetten, M.S.M., and Lüke, C., Stratification of diversity and activity of methanogenic and methanotrophic microorganisms in a nitrogen-fertilized Italian paddy soil, Front. Microbiol., 2017, vol. 8, pp. 1–15.
van der Ha, D., Nachtergaele, L., Kerckhof, F.M., Rameiyanti, D., Bossier, P., Verstraete, W., and Boon, N., Conversion of biogas to bioproducts by algae and methane oxidizing bacteria, Environ. Sci. Technol., 2012, vol. 46, pp. 13425–13431.
Weisburg, W.G., Barns, S.M., Pelletier, D.A., and Lane D.J., 16S ribosomal DNA amplification for phylogenetic study, J. Bacteriol., 1991, vol. 173, pp. 697–703.
Yilmaz, P., Parfrey, L.W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., Schweer, T., Peplies, J., Ludwig, W., and Glöckner, F.O., The SILVA and “all-species Living Tree Project (LTP)” taxonomic frameworks, Nucleic Acids Res., 2014, vol. 42, pp. 643–648.
Funding
The work was supported by the Russian Federation Ministry of Science and Higher Education.
Author information
Authors and Affiliations
Contributions
SND and NVP proposed the outline of the work. TVM and AIM collected the samples and determined their physicochemical parameters. IYuO and OVD carried out molecular analysis of microbial diversity and isolation and identification of methanotrophic strains. RZS determined the growth characteristics of the isolates. IYuO, NVP, and SND wrote the text of the article. All authors participated in discussion of the results.
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by P. Sigalevich
Rights and permissions
About this article
Cite this article
Oshkin, I.Y., Danilova, O.V., Suleimanov, R.Z. et al. Thermotolerant Methanotrophic Bacteria from Sediments of the River Chernaya, Crimea, and Assessment of Their Growth Characteristics. Microbiology 90, 588–597 (2021). https://doi.org/10.1134/S0026261721050131
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026261721050131