Abstract—
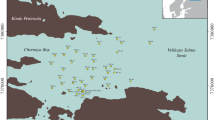
Recently discovered fields of seeps in western Siberia are an important, previously unknown source of methane. Seeps are located in the floodplains of small rivers and vary in shape, size and localization. Methane fluxes from seeps may be high, making them the sources of high regional importance. However, the data on the seep bacterial community composition are scarce, concentrating mainly on methanotrophic bacteria. In the present work, the overall bacterial diversity in the sediments of shallow-water seeps at the floodplain of the Bolshaya Rechka River was studied using high-throughput 16S rRNA gene sequencing. Molecular analysis revealed that Gammaproteobacteria and Actinobacteria were the most abundant bacterial groups (28.5–33.8 and 11–13.2% of total 16S rRNA genes, respectively). A significant part of the sequences belonged to Chloroflexi, Desulfobacterota, and Bacteroidota. Acidobacteriota and Verrucomicrobiota were responsible for 1.5–2.7 and 1.0–1.9%, respectively. The methanotrophic community was dominated by bacteria of the genus Methylobacter. The most numerous species-level OTU (8% of all 16S rRNA gene sequences) belonged to Methylobacter tundripaludum (97% identity). Methanotrophic Alphaproteobacteria were not detected in the seeps. Our results confirm the presence of an active bacterial community in the sediments of shallow-water seeps, with predominance of methanotrophic Gammaproteobacteria.





Similar content being viewed by others
REFERENCES
Belova, S.E., Oshkin, I.Y., Dedysh, S.N., Glagolev, M.V., Lapshina, E.D., and Maksyutov, S.S., Methanotrophic bacteria in cold seeps of the floodplains of northern rivers, Microbiology (Moscow), 2013, vol. 82, pp. 743–750.
Bolyen, E., Rideout, J.R., Dillon, M.R., Bokulich, N.A., Abnet, C.C., Al-Ghalith, G.A., Alexander, H., Alm, E.J., Arumugam, M., Asnicar, F., Bai, Y., Bisanz, J.E., Bittinger, K., Brejnrod, A., Brislawn, C.J., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2, Nat. Biotechnol., 2019, vol. 37, pp. 852–857.
Bourne, D.G., McDonald, I.R., and Murrell, J.C., Comparison of pmoA PCR primer sets as tools for investigating methanotroph diversity in three Danish soils, Appl. Environ. Microbiol., 2001, vol. 67, pp. 3802–3809.
Bousquet, P., Ciais, P., Miller, J.B., Dlugokencky, E.J., Hauglustaine, D.A., Prigent, C., van der Werf, G.R., Peylin, P., Brunke, E.-G., Carouge, C., Langenfelds, R.L., Lathière, J., Papa, F., Ramonet, M., Schmidt, M., et al., Contribution of anthropogenic and natural sources to atmospheric methane variability, Nature, 2006, vol. 443, pp. 439–443.
Callahan, B.J., McMurdie, P.J., Rosen, M.J., Han, A.W., Johnson, A.J.A., and Holmes, S.P., DADA2: high-resolution sample inference from Illumina amplicon data, Nat. Methods, 2016, vol. 13, pp. 581–583.
Ciais, P., Sabine, C., Bala, G., Bopp, L., Brovkin, V., Canadell, J., Chhabra, A., DeFries, R., Galloway, J., Heimann, M. Jones, C., Le Quéré, C., Myneni, R.B., Piao, S., and Thornton, P., Carbon and other biogeochemical cycles, in Climate Change 2013: The Physical Science Basis, Contribution of Working Group I to the Fifth Assessment Report of IPCC, Stocker, T.F., Qin, D., Plattner, G.-K., Tignor, M., Allen, S.K., Boschung, J., Nauels, A., Xia, Y., Bex, V., and Midgley, P.M., Eds., Cambridge: Cambridge Univ. Press, 2013, pp. 465–570.
da Rocha, U.N., Plugge, C.M., George, I., van Elsas, J.D., and van Overbeek, L.S., The rhizosphere selects for particular groups of Acidobacteria and Verrucomicrobia, PLoS One, 2013, vol. 8, art. e82443.
Fadrosh, D., Ma, B., Gajer, P., Sengamalay, N., Ott, S., Brotman, R., and Ravel, J., An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform, Microbiome, 2014, vol. 2, p. 6.
Fletcher, S.E.M. and Schaefer, H., Rising methane: a new climate challenge, Science, 2019, vol. 364, pp. 932–933.
Holmes, A.J., Costello, A., Lidstrom, M.E., and Murrell, J.C., Evidence that particulate methane monooxygenase and ammonium monooxygenase may be evolutionarily related, FEMS Microbiol. Lett., 1995, vol. 132, pp. 203–208.
Hope, D., Dawson, J.J., Cresser, M.S., and Billett, M.F., A method for measuring free CO2 in upland streamwater using headspace analysis, J. Hydrol., 1995, vol. 166, pp. 1–14.
Huber, K.J., Geppert, A.M., Wanner, G., Fösel, B.U., Wüst, P.K., and Overmann, J., The first representative of the globally widespread subdivision 6 Acidobacteria, Vicinamibacter silvestris gen. nov., sp. nov., isolated from subtropical savannah soil, Int. J. Syst. Evol. Microbiol., 2016, vol. 66, pp. 2971–2979.
Hugerth, L.W., Wefer, H.A., Lundin, S., Jakobsson, H.E., Lindberg, M., Rodin, S. Engstrand, L., and Andersson, A.F., DegePrime, a program for degenerate primer design for broad-taxonomic-range PCR in microbial ecology studies, Appl. Environ. Microbiol., 2017, vol. 80, pp. 5116–5123.
Janssen, P.H., Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes, Appl. Environ. Microbiol., 2006, vol. 72, pp. 1719–1728.
Kalyuzhnaya, M.G., Lapidus, A., Ivanova, N., Copeland, A.C., McHardy, A.C., Szeto, E., Salamov, A., Grigoriev, I.V., Suciu, D., Levine, S.R., Markowitz, V.M., Rigoutsos, I., Tringe, S.G., Bruce, D.C., Richardson, P.M., et al., High-resolution metagenomics targets specific functional types in complex microbial communities, Nat. Biotechnol., 2008, vol. 26, pp. 1029–1034.
Kielak, A.M., Barreto, C.C., Kowalchuk, G.A., van Veen, J.A., and Kuramae, E.E., The ecology of Acidobacteria: moving beyond genes and genomes, Front. Microbiol., 2016, vol. 7, art. 744.
Kim, M., Pak, S., Rim, S., Ren, L., Jiang, F., Chang, X., Liu, P., Zhang, Y., Fang, C., Zheng, C., and Peng, F., Luteolibacter arcticus sp. nov., isolated from high Arctic tundra soil, and emended description of the genus Luteolibacter, Int. J. Syst. Evol. Microbiol., 2015, vol. 65, pp. 1922–1928.
Kubista, M., Andrade, J.M., Bengtsson, M., Forootan, A., Jonák, J., Lind, K., Sindelka, R., Sjöback, R., Sjögreen, B., Strömbom, L., Ståhlbergag, A., and Zorica, N., The real-time polymerase chain reaction, Mol. Aspects Med., 2006, vol. 2, pp. 95–125.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K., MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms, Mol. Biol. Evol., 2018, vol. 35, pp. 1547–1549.
Liao, L., Xu, X.W., Jiang, X.W., Wang, C.S., Zhang, D.S., Ni, J-Y., and Wu, M., Microbial diversity in deep-sea sediment from the cobalt-rich crust deposit region in the Pacific Ocean, FEMS Microbiol. Ecol., 2011, vol. 78, pp. 565–585.
Lin, J.Y., Russell, J.A., Sanders, J.G., and Wertz, J.T., Cephaloticoccus gen. nov., a new genus of “Verrucomicrobia” containing two novel species isolated from Cephalotes ant guts, Int. J. Syst. Evol. Microbiol., 2016, vol. 66, pp. 3034–3040.
Merkel, A.Y., Podosokorskaya, O.A., Toshchakov, S.V., and Tarnovetskii, I.Y., Analysis of 16S rRNA primer systems for profiling of thermophilic microbial communities, Microbiology (Moscow), 2019, vol. 88, pp. 671–680.
Navarrete, A.A., Kuramae, E.E., de Hollander, M., Pijl, A.S., van Veen, J.A., and Tsai, S.M., Acidobacterial community responses to agricultural management of soybean in Amazon forest soils, FEMS Microbiol. Ecol., 2013, vol. 83, pp. 607–621.
Onley, J.R., Ahsan, S., Sanford, R.A., and Löffler, F.E., Denitrification by Anaeromyxobacter dehalogenans, a common soil bacterium lacking the nitrite reductase genes nirS and nirK, Appl. Environ Microbiol., 2017, vol. 84, art. e01985-17.
Oshkin, I.Y., Belova, S.E., Danilova, O.V., Miroshnikov, K.K., Rijpstra, W.I.C., Sinninghe Damste, J.S., Liesack, W., and Dedysh, S.N., Methylovulum psychrotolerans sp. nov., a cold-adapted methanotroph from low-temperature terrestrial environments, and emended description of the genus Methylovulum, Int. J. Syst. Evol. Microbiol., 2016, vol. 66, pp. 2417–2423.
Oshkin, I.Y., Wegner, C.E., Lüke, C., Glagolev, M.V., Filippov, I.V., Pimenov, N.V., Liesack, W., and Dedysh, S.N., Gammaproteobacterial methanotrophs dominate cold methane seeps in floodplains of West Siberian rivers, Appl. Environ. Microbiol., 2014, vol. 80, pp. 5944–5954.
Oswald, K., Graf, J., Littmann, S., Tienken, D., Brand, A., Wehrli, B., Albertsen, M., Daims, H., Wagner, M., Kuypers, M., Schubert, C.J., and Milucka, J., Crenothrix are major methane consumers in stratified lakes, ISME J., 2017, vol. 11, pp. 2124–2140.
Pascual, J., Garcia-Lopez, M., Gonzalez, I., and Genilloud, O., Luteolibacter gellanilyticus sp. nov., a gellan-gum-degrading bacterium of the phylum Verrucomicrobia isolated from miniaturized diffusion chambers, Int. J. Syst. Evol. Microbiol., 2017, vol. 67, pp. 3951–3959.
Pascual, J., Wüst, P.K., Geppert, A., Foesel, B.U., Huber, K.J., and Overmann, J., Novel isolates double the number of chemotrophic species and allow the first description of higher taxa in Acidobacteria Subdivision 4, Syst. Appl. Microbiol., 2015, vol. 38, pp. 534–544.
Qiu, Y.L., Kuang, X.Z., Shi, X.S., Yuan, X.Z., and Guo, R.B., Terrimicrobium sacchariphilum gen. nov., sp. nov., an anaerobic bacterium of the class “Spartobacteria” in the phylum Verrucomicrobia, isolated from a rice paddy field, Int. J. Syst. Evol Microbiol., 2014, vol. 64, pp. 1718–1723.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., Peplies, J., and Glöckner, F.O., The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools, Nucl. Acids Res., 2013, vol. 41, pp. 590–596.
Ratcliffe, J.L., Creevy, A., Andersen, R., Zarov, E., Gaffney, P.P., Taggart, M.A., Mazeie, Y., Tsyganov, A.N., Rowson, J.G., Lapshina, E.D., and Payne, R.J., Ecological and environmental transition across the forested-to-open bog ecotone in a west Siberian peatland, Sci. Total Environ., 2017, vol. 607, pp. 816–828.
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F., VSEARCH: a versatile open source tool for metagenomics, PeerJ., 2016, vol. 4, art. e2584.
Sabrekov, A.F., Terentieva, I.E., Glagolev, M.V., Litti, Y.V., and Maksyutov, S.S., Methane seeps in West Siberian middle taiga river floodplains: origin and fluxes, AGU Fall Meeting, 2020.
Sanford, R.A., Wagner, D.D., Wu, Q., Chee-Sanford, J.C., Thomas, S.H., Cruz-García, C., Rodríguez, G., Massol-Deyá, A., Krishnani, K.K., Ritalahti, K.M., Nissen, S., Konstantinidis, K.T., and Löffler, F.E. Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils, Proc. Natl. Acad. Sci. U. S. A., 2012, vol. 109, pp. 19709–19714.
Terentieva, I.E., Glagolev, M.V., Lapshina, E.D., Sabrekov, A.F., and Maksyutov, S., Mapping of West Siberian taiga wetland complexes using Landsat imagery: implications for methane emissions, Biogeosci., 2016, vol. 13, pp. 4615–4626.
Vieira, S., Luckner, M., Wanner, G., and Overmann, J., Luteitalea pratensis gen. nov., sp. nov. a new member of subdivision 6 Acidobacteria isolated from temperate grassland soil, Int. J. Syst. Evol. Microbiol., 2017, vol. 67, pp. 1408–1414.
Yilmaz, P., Parfrey, L.W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., Schweer, T., Peplies, J., Ludwig, W., and Glöckner, F.O., The SILVA and “all-species Living Tree Project (LTP)” taxonomic frameworks, Nucl. Acids Res., 2014, vol. 42, pp. 643–648.
Yoon, J., Matsuo, Y., Adachi, K., Nozawa, M., Matsuda, S., Kasai, H., and Yokota, A., Description of Persicirhabdus sediminis gen. nov., sp. nov., Roseibacillus ishigakijimensis gen. nov., sp. nov., Roseibacillus ponti sp. nov., Roseibacillus persicicus sp. nov., Luteolibacter pohnpeiensis gen. nov., sp. nov. and Luteolibacter algae sp. nov., six marine members of the phylum “Verrucomicrobia”, and emended descriptions of the class Verrucomicrobiae, the order Verrucomicrobiales and the family Verrucomicrobiaceae, Int. J. Syst. Evol. Microbiol., 2008, vol. 58, pp. 998–1007.
Funding
The study was supported by the Russian Science Foundation, project no. 19-77-10074. The logistics of the work was organized with the support of a grant from the Government of the Tyumen region in accordance with the program of the West Siberian Interregional Scientific and Educational Center of the world level within the framework of the national project “Science.”
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by E. Babchenko
Rights and permissions
About this article
Cite this article
Danilova, O.V., Ivanova, A.A., Terent’eva, I.E. et al. Microbial Community Composition of Floodplains Shallow-Water Seeps in the Bolshaya Rechka Floodplain, Western Siberia. Microbiology 90, 632–642 (2021). https://doi.org/10.1134/S0026261721050040
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026261721050040