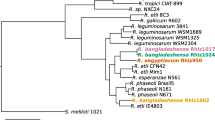
Abstract
The theoretical and experimental data on salt tolerance of root nodule bacteria Sinorhizobium meliloti (Ensifer meliloti), an alfalfa symbiont, and on genetic determination of this feature are reviewed. Extensive data are provided on the genes affecting adaptation of proteobacteria and on the groups of genes with activity depending on the osmolarity of the medium. Structural and functional polymorphism of the bet genes involved in betaine synthesis and transport in S. meliloti is discussed. The phenotypic and genotypic polymorphism in 282 native rhizobial strains isolated from the centers of alfalfa diversity affected by aridity and salinity is discussed. The isolates from the Aral Sea area and northern Caucasus were shown to possess the betC gene represented by two types of alleles: the dominant Atype allele found in Rm1021 and the less common divergent Etype allele, which was revealed in regions at the frequencies of 0.35 and 0.48, respectively. In the isolates with the salt-tolerant phenotype, which were isolated from root nodules and subsequently formed less effective symbioses with alfalfa, the frequency of Etype alleles was 2.5 times higher. Analysis of the nucleotide and amino acid sequences of the Etype allele of the betC gene revealed that establishment of this allele in the population was a result of positive selection. It is concluded that diversification of the functionally diverse bet genes occurring in S. meliloti affects the salt tolerance and symbiotic effectiveness of rhizobia.
Similar content being viewed by others
References
Flowers, T.J., Improving crop salt tolerance, J. Experim. Bot., 2004, vol. 55, no. 396, pp. 307–319.
FAO. Food outlook. Global Market Analysis, 2007. http://www.fao.Food outlook.com
Souss, M., Santamaria, M., Ocana, A., and Lluch, C., Effects of salinity on protein and lipopolysaccharide pattern in a salttolerant strain of Mesorhizobium ciceri, J. Appl. Microbiol., 2001, vol. 90, no. 3, pp. 476–481.
Zahran, H.H., Rhizobiumlegume symbiosis and nitro gen fixation under severe conditions and in an arid cli mate, Microbiol. Mol. Biol. Rev., 1999, vol. 63, no. 4, pp. 968–989.
Dzyubenko, N.I., Chapurin, V.F., Bukhteeva, A.V., and Soskov, Yu.D., Mobilization and investigation of perennial fodder crops in view of N. Vavilov's heritage, Tr. Prikl. Bot. Gen. Select., 2007, vol. 164, pp. 153–163.
Lesins, K. and Lesins, I., Genus Medicago (Legumino sae), a taxogenetic study, Plant Ecol., 1982, pp. 50–92.
Small, E., Pollenovule patterns in tribe Trifolieae (Leguminosae), Plant Syst. Evol., 1988, vol. 160, nos. Iss. 3–4, pp. 195–205.
Shamsutdinov, Z.Sh., Methods for ecological restora tion of arid systems in the regions of grazing livestock sector, Stepnoi Byull. Novosib. Univ., 2002, vol. 11, pp. 21–26.
Stepanova, G.V., Muntyan, V.S., and Rumyantseva, M.L., Response of the new Agniya alfalfa variety to inoculation with root nodule bacteria, Adaptivnoe kormoproizvodstvo, 2013, no. 3(15), pp. 43–48. (Adaptive Forage Produc tion). http://www. adaptagro.ru
Kulkarni, S., Surange, S., and Nautiyal, C.S., Crossing the limits of Rhizobium existence in extreme condi tions, Curr. Microbiol., 2000, vol. 41, pp. 402–409.
Silva, C., Kan, F.L., and MartinezRomero, E., Popu lation genetic structure of Sinorhizobium meliloti and S. medicae isolated from nodules of Medicago spp. in Mexico, FEMS Microbiol. Ecol., 2007, vol. 60, pp. 477–489.
Eardly, B.D., Materon, L.A., Smith, N.H., Johnson, D.A., Rumbaugh, M.D., and Selander, R.K., Genetic structure of natural populations of the nitrogen fixing bacterium Rhizobium meliloti, Appl. Environ. Microbiol., 1990, vol. 56, pp. 187–194.
Wails, R.J., Wells, D.H., and Long, S.R., Analysis of differences between Sinorhizobium meliloti 1021 and 2011 strains using the host calcium spiking response, Mol. Plant–Microbe Interact., 2002, vol. 15, no. 12, pp. 1245–1252.
Elboutahiri, N., ThamiAlami, I., and Udupa, S.M., Phenotypic and genetic diversity in Sinorhizobium meliloti and S. medicae from drought and salt affected regions of Morocco, BMC Microbiol., 2010, vol. 10, p. 15.
Rangin, C., Brunel, B., CleyetMarel, J.C., Per rineau, M.M., and Bena, G., Effects of Medicago trun catula genetic diversity, rhizobial competition, and strain effectiveness on the diversity of a natural Sinorhizobium species community, Appl. Environ. Microbiol., 2008, vol. 74, no. 18, pp. 5653–5661.
Andronov, E.E., Rumiantseva, M.L., and Simarov, B.V., Genetic diversity of a natural population of Sinorhizo bium meliloti, detected during analysis of a cryptic plas mid and ISRm20112 fingerprints, Russ. J. Genet., 2001, vol. 37, no. 5, pp. 610–616.
GubryRangin, C., Garcia, M., and Bena, G., Partner choice in Medicago truncatula–Sinorhizobium symbio sis, Proc. Biol. Sci., 2010, vol. 277, no. 1690, pp. 1947–1951.
Ibragimova, M.V., Rumiantseva, M.L., Onishchuk, O.P., Belova, V.S., Kurchak, O.N., Andronov, E.E., Dzyu benko, N.I., and Simarov, B.V., Symbiosis between the nodule bacterium Sinorhizobium meliloti and alfalfa (Medicago sativa) under salinization conditions, Microbiology (Moscow), 2006, vol. 75, no. 1, pp. 77–81.
Yurkov, A.P., Yakobi, L.M., Roumiantseva, M.L., and Stepanova, G.V., Capacity of alfalfa MIS1 line mutants for symbiosis formation with root nodule bac teria, Estestv. Tekhn. Nauki, 2012, no. 6, pp. 124–128.
Simarov, B.V. and Roumiantseva, M.L., Genetic approaches for establishing highly efficient effective, competitive symbioses between root nodule bacteria and alfalfa under impact of abiotic stress factors (salin ization), in Mat. Konf. “Orientirovannye fundamen tal’nye issledovaniya i ikh realizatsiya v APK Rossii” (Oriented Basic Research and Their Realization in the Russian AgroIndustrial Complex), St. Petersburg, 2008, pp.11–12.
Zahran, H.H., Rhizobia from wild legumes: diversity, taxonomy, ecology, nitrogen fixation and biotechnol ogy, J. Biotechnol., 2001, vol. 91. nos. 2–3. pp. 143–153.
Abdelmoumen, H., FilaliMaltouf, A., Neyra, M., Belabed, A., Missbah, El., and Idrissi, M., Effect of high salts concentrations on the growth of rhizobia and responses to added osmotica, J. Appl. Microbiol., 1999, vol. 86, pp. 889–898.
Bena, G., Lyet, A., Huguet, T., and Olivieri, I., Medicago–Sinorhizobium symbiotic specificity evolution and the geographic expansion of Medicago, J. Evol. Biol., 2005, vol. 18, no. 6, pp. 1547–1558.
Wei, W., Jiang, J., Li, X., Wang, L., and Yang, S.S., Iso lation of saltsensitive mutants from Sinorhizobium meliloti and characterization of genes involved in salt tolerance, Lett. Appl. Microbiol., 2004, vol. 39, pp. 278–283.
Roumiantseva, M.L., Belova, V.S., Onishchuk, O.P., Andronov, E.E., Kurchak, O.N., Chizhevskaya, E.P., Rumyantseva, T.B., and Simarov, B.V., Polymorphism of the bet genes in Sinorhizobium meliloti strains from alfalfa genetic centers, Sel’skokhoz. Biol., 2011, no. 3, pp. 48–54.
Dogra, T., Priyadarshini, A., Kumar, A., and Kumar Singh, N., Identification of genes involved in salt tolerance and symbiotic nitrogen fixation in chick pea rhizobium Mesorhizobium ciceri Ca181, Symbiosis, 2013, vol. 61, pp. 135–143.
Aguilar, O.M., Riva, O., and Peltzer, E., Analysis of Rhizobium etli and of its symbiosis with wild Phaseolus vulgaris supports coevolution in centers of host diversi fication, Proc. Natl. Acad. Sci. U. S. A., 2004, vol. 101, no. 37, pp. 13548–13553.
Roumiantseva, M.L. Genetic resources of nodule bac teria (review), Russ. J. Genet., 2009, vol. 45, no. 9, pp. 1013–1026.
Vavilov, N.I., Centers of origin of cultivated plants, in Trudy po prikladnoi botanike, genetike i selektsii (Works on Applied Botany, Genetics, and Selection), Lenin grad: Tip. im. Gutenberga, 1926, vol. 16, no. 2, pp. 3–248.
Sinskaya, E.N., Ecological system for selection of fod der plants, in Prilozh. №62k Tr. po prikl. bot., gen. i sel (Suppl. no. 62 to Proc. Appl. Bot., Genet., Selct.), Leningrad: VIR, 1933.
Kumar, H., Arora, N.K., Kumar, V., and Maheshwari, D.K., Isolation, characterization and selection of salttolerantrhizobia nodulating Acacia catechu and Acacia nilotica, Symbiosis, 1999, vol. 26, pp. 279–288.
Boncompagni, E. and Poggi, M.C., and Le Rudulier, D., Occurrence of choline and glycine betaine uptake and metabolism in the family Rhizobiaceae and their roles in osmoprotection, Appl. Environ. Microbiol., 1999, vol. 65, pp. 2072–2077.
Boscari, A. Van de Sype, G., Le Rudulier, D., and Mandon, K., Overexpression of BetS, a Sinorhizobium meliloti highaffinity betaine transporter, in bacteroids from Medicago sativa nodules sustains nitrogen fixation during early salt stress adaptation, Mol. Plant–Microbe Interact., 2006, vol. 19, no. 8, pp. 896–903.
Roumyantseva, M.L., Andronov, E.E., Onichtchouk, O.P., Kurchak, O.N., Ibragimova, M.V., Dzubenko, N.I., Lindstroem, K., Priefer, U.B., Giuntini, E., Bazzicalupo, M., and Simarov, B.V., Sinorhizo bium isolates from saltaffected Aral Sea asin, 5th Eur. Nitrogen Fixation Conf., Norwich, 2002.
Young, J.P.W., Crossman, L.C., Johnston, A.W.B., Thomson, N.R., Ghazoui, Z.F., Hull, K.H., Wexler, M., Curson, A.R.J., Todd, J.D., Poole, P.S., Mauchline, T.H., East, A.K., Quail, M.A., Churcher, C., Arrowsmith, C., Cherevach, I., Chilling worth, T., Clarke, K., Cronin, A., Davis, P., Fraser, A., Hance, Z., Hauser, H., Jagels, K., Moule, S., Mungall, K., Norbertczak, H., Rabbinowitsch, E., Sanders, M., Simmonds, M., Whitehead, S., and Parkhill, J., The genome of Rhizobium leguminosarum has recognizable core and accessory components, Genome Biol., 2006, 7R34. doi: 10.1186/gb-2006-7-4-r34
Roumiantseva, M.L., Simarov, B.V., Onishchuk, O.P., Andronov, E.E., Chizhevskaya, E.P., Belova, V.S., Kurchak, O.N., Muntyan, A.N., Rumyantseva, T.B., and Zatovskaya, T.V., Biologicheskoe raznoobrazie kluben’kovykh bakterii v ekosistemakh i agrotsenoz. Teoreticheskie osnovy i metody (Biological Diversity of Root Nodule Bacteria in Ecosystems and Agrocenoses. Theoretical Foundations and Methods), Roumiant-seva, M.L. and Simarov B.V., Eds., S.-Pb.–Pushkin: VNIISKhM, 2011.
Onishchuk, O.P., Roumiantseva, M.L., Provorov, N.A., and Simarov, B.V., Variability of Sinorhizobium meliloti strains in characteristics determining their saprophytic survival under salinization conditions, Sel’skokhoz. Biol., 2009, no. 1, pp. 77–82.
Roumiantseva, M.L., Onischuk, O.P., Belova, V.S., Kurchak, O.N., and Simarov B.V., Polymorphism of Sinorhizobium meliloti strains isolated from diversity centers of alfalfa in various soil and climatic conditions, Rus. J. Genet.: Appl. Res., 2011, vol. 1, no. 2, pp. 97–102.
Alloing, G., Travers, I., and Sagot, B., Le Rudulier, D., and Dupont, L., Proline betaine uptake in Sinorhizo bium meliloti: characterization of Prb, an Opplike ABC transporter regulated by both proline betaine and salinity stress, J. Bacteriol., 2006, vol. 188, no. 17, pp. 6308–6317.
Galibert, F., Finan, T.M., Long, S.R., Puehler, A., Abola, P., Ampe, F., BarloyHubler, F., Barnett, M.J., Becker, A., Boistard, P., Bothe, G., Boutry, M., Bowser, L., Buhrmester, J., Cadieu, E., Capela, D., Chain, P., Cowie, A., Davis, R.W., Dreano, S., Federspiel, N.A., Fisher, R.F., Gloux, S., Godrie, T., Goffeau, A., Golding, B., Gouzy, J., Gurjal, M., HernandezLucas, I., Hong, A., Huizar, L., Hyman, R.W., Jones, T., Kahn, D., Kahn, M.L., Kalman, S., Keating, D.H., Kiss, E., Komp, C., Lelaure, V., Masuy, D., Palm, C., Peck, M.C., Pohl, T.M., Portetelle, D., Purnelle, B., Ramsperger, U., Surzycki, R., Thebault, P., Vandenbol, M., Vorholter, F.J., Weidner, S., Wells, D.H., Wong, K., Yeh, K.C., and Batut, J., The composite genome of the legume sym biont Sinorhizobium meliloti, Science, 2001, vol. 293, no. 5530, pp. 668–672.
Sun, S., Guo, H., and Xu, J., Multiple gene genealogi cal analyses reveal both common and distinct popula tion genetic patterns among replicons in the nitrogen fixing bacterium Sinorhizobium meliloti, Microbiology (UK), 2006, vol. 152, pp. 3245–3259.
Guo, H., Sun, S., Finan, T.M., and Xu, J., Novel DNA sequences from natural strains of the nitrogenfixing symbiotic bacterium Sinorhizobium meliloti, Appl. Environ. Microbiol., 2005, vol. 71, no. 11, pp. 7130–7138.
Schwedock, J.S. and Long, S.R., Rhizobium meliloti genes involved in sulfate activation: the two copies of nodPQ and a new locus, saa, Genetics, 1992, vol. 132, no. 4, pp. 899–909.
Reguera, M., Lloret, J., Margaret, I., Vinardell, J.M., Martin, M., Buendia, A., Rivilla, R., RuizSainz, J.E., Bonilla, I., and Bolanos, L., Gene Smb21071 of plas mid pSymB is required for osmoadaptation of Sinorhizobium meliloti 1021 and is implicated in modi fications of cell surface polysaccharides structure in response to hyperosmotic stress, Can. J. Microbiol., 2009, vol. 10, pp. 1145–1152.
Stiens, M., Schneiker, S., Keller, M., Kuhn, S., Puehler, A., and Schlueter, A., Sequence analysis of the 144kilobase accessory plasmid pSmeSM11a, isolated from a dominant Sinorhizobium meliloti strain identi fied during a longterm field release experiment, Appl. Environ. Microbiol., 2006, vol. 72, pp. 3662–3672.
Stiens, M., Schneiker, S., Puehler, A., and Schlueter, A., Sequence analysis of the 181kb accessory plasmid pSmeSM11b, isolated from a dominant Sinorhizobium meliloti strain identified during a longterm field release experiment, FEMS Microbiol. Lett., 2007, vol. 271, pp. 297–309.
Roumiantseva, M.L., Andronov, E.E., Sagulenko, V.V. Onishuk, O.P., Provorov, N.A., and Simarov, B.V., Plasmids in strain Sinorhizobium meliloti P108 in the course of symbiosis with alfalfa Medicago sativa, Russ. J. Genet., 2004, vol. 40, no. 4, pp. 356–362.
Ermilova, E.V., Molekulyarnye aspekty adaptatsii prokariot (Molecular Aspects of Prokaryotic Adapta tion), St. Petersburg: Khimizdat, 2012.
McIntyre, H.J., Davies, H., Hore, T.A., Miller, S.H., Dufour, J.P., and Ronson, C.W., Trehalose biosynthesis in Rhizobium leguminosarum bv. trifolii and its role in desiccaion tolerance, Appl. Environ. Microbiol., 2007, vol. 73, no. 12, pp. 3984–3992.
Vriezen, J.A.C., de Bruijn, F.J., and Nuesslein, K., Desiccation responses and survival of Sinorhizobium meliloti USDA 1021 in relation to growth phase, tem perature, chloride and sulfate availability, Lett. Appl. Microbiol., 2006, vol. 42, no. 2, pp. 172–178.
Becker, A., Schmidt, M., Jaeger, W., and Puehler, A., New gentamicinresistance and lacZ promoterprobe cassettes suitable for insertion mutagenesis and genera tion of transcriptional fusions, Gene, 1995, vol. 162, pp. 37–39.
Vinuesa, P., NeumannSilkow, F., PaciosBras, C., Spaink, H.P., MartinezRomero, E., and Werner, D., Genetic analysis of a pHregulated operon from Rhizo bium tropici CIAT899 involved in acid tolerance and nodulation competitiveness, Mol. Plant–Microbe Inter act., 2003, vol. 16, no. 2, pp. 159–168.
Wei, W., Gu, Z.J., Zhang, B., Wang, L., and Yang, S.S., Cloning and complementation analysis of greA gene involved in salt tolerance of Sinorhizobium meliloti, Ann. Microbiol., 2007, vol. 57, no. 2, pp. 289–291.
Jiang, J.Q., Wei, W., Du, B.H., Li, X.H., Wang, L., and Yang, S.S., Salttolerance genes involved in cation efflux and osmoregulation of Sinorhizobium fredii RT19 detected by isolation and characterization of Tn5 mutants, FEMS Microbiol. Lett., 2004, vol. 239, no. 1, pp. 139–146.
Chizhevskaya, E.P., Onishchuk, O.P., Andronov, E.E., and Simarov, B.V., Application of sitedirected mutagenesis for investigation of the function of the SMb20332 gene in root nodule bacteria Sinorhizobium meliloti, Sel’skokhoz. Biol., 2011, no. 3, pp. 55–61.
Belova, V.S., Yurgel’, S.N., Rais, D., Rumyantseva, T.B., Simarov, B.V., and Roumiantseva, M.L., Survival of Sinorhizobium meliloti strain CIAM1775 in soils of differ ent acidity, in Materialy Vserossiiskoi nauchnoprak ticheskoi konferentsii “Perspektivnye napravleniya issledo vanii v zemledelii i rastenievodstve” UlGTU (Proc. AllRuss. Sci.Pract. Conf. “Prospective Directions of Research in Agriculture and Crop Sector), Ul’yanovsk, 2011, pp. 35–39.
Ohwada, T., Sasaki, Y., Koike, H., Igawa, K., and Sato, T., Correlation between NaCl sensitivity of Rhizobium bacteria and ineffective nodulation of legu minous plants, Boisci. Biotechnol. Biochem., 1998, vol. 62, no. 11, pp. 2086–2090.
Chien, C.T., Maundu, J., Cavaness, J., Dandurand, L.M., and Orser, C.S., Characterization of salt tolerant and saltsensitive mutants of Rhizobium legu minosarum bv. viciae strain C12046, FEMS Microbiol. Lett., 1992, vol. 90, pp. 135–140.
Pobigaylo, N., Szymczak, S., Nattkemper, T.W., and Becker, A., Identification of genes relevant to symbiosis and competitiveness in Sinorhizobium meliloti using sig naturetagged mutants, Mol. Plant–Microbe Interact., 2008, vol. 21, no. 2, pp. 219–231.
Pobigaylo, N., Wetter, D., Szymczak, S., Schiller, U., Kurtz, S., Meyer, F., Nattkemper, T.W., and Becker, A., Construction of a large signaturetagged miniTn5 transposon library and its application to mutagenesis of Sinorhizobium meliloti, Appl. Environ. Microbiol., 2006, vol. 72, no. 6, pp. 4329–4337.
GuaschVidal, B., Brussel, A.A.N., Estevez, J., Bellogin, R., Ollero, F.J., Espuny, M.R., and Megias, M., Nod factor production and abiotic stress in Rhizobium, in Beneficial PlantMicrobial Interactions. Ecology and Applications, Belen Rodelas Gonzalez, M. and Gonza lezLopez, J., Eds., New York: CRC, 2013, pp. 71–98.
DominguezFerreras, A., PerezArnedo, R., Becker, A., Olivares, J., Soto, M.J., and Sanjuan, J., Transcriptome profiling reveals the importance of plasmid pSymB for osmoadaptation of Sinorhizobium meliloti, J. Bacteriol., 2006, vol. 188, pp. 7617–7626.
Boscari, A., Mandon, K., Dupont, L., and Poggi, M.C., and Le Rudulier, D., BetS is a major glycine betaine/pro line betaine transporter required for early osmotic adjust ment in Sinorhizobium meliloti, J. Bacteriol., 2002, vol. 184, no. 10, pp. 2654–2663.
Angelidis, A.S. and Smith, G.M., Role of the glycine betaine and carnitine transporters in adaptation of Listeria monocytogenes to chill stress in defined medium, Appl. Environ. Microbiol., 2003, vol. 69, no. 12, pp. 7492–7498.
Davidson, A.L., Dassa, E., Orelle, C., and Chen, J., Structure, function, and evolution of bacterial ATP binding cassette systems, Microbiol. Mol. Biol. Rev., 2008, vol. 72, pp. 317–364.
Pumirat, P., Cuccui, J., Stabler, R.A., Stevens, J.M., Muangsombut, V., Singsuksawat, E., Stevens, M.P., Wren, B.W., and Korbsrisate, S., Global transcriptional profiling of Burkholderia pseudomallei under salt stress reveals differential effects on the Bsa type III secretion system, BMC Microbiol., 2010, vol. 14. doi: 10.1186/1471218010171
Sagot, B., Gaysinski, M., Mehiri, M., Guigonis, J.M., Le Rudulier, D., and Alloing, G., Osmotically induced synthesis of the dipeptide Nacetylglutaminyl glutamine amide is mediated by a new pathway con served among bacteria, Proc. Natl. Acad. Sci. U. S. A., 2010, vol. 107, no. 28, pp. 12652–12657.
Rueberg, S., Tian, Z.X., Krol, E., Linke, B., Meyer, F., Wang, Y., Puehler, A., Weidner, S., and Becker, A., Con struction and validation of a Sinorhizobium meliloti whole genome DNA microarray: genomewide profiling of osmoadaptive gene expression, J. Biotechnol., 2003, vol. 106, pp. 255–268.
Finan, T.M., Weidner, S., Wong, K., Buhrmester, J., Chain, P., Vorhoelter, F.J., HernandezLucas, I., Becker, A., Cowie, A., Gouzy, J., Golding, B., and Puehler, A., The complete sequence of the 1.683kb pSymB megaplasmid from the N2fixing endosymbiont Sinorhizobium meliloti, Proc. Natl. Acad. Sci. USA, 2001, vol. 98, no. 17, pp. 9889–9894.
Muntyan, V.S. and Roumiantseva, M.L., Core genome of stressresistant Sinorhizobium spp. strains, inoculants of economically important leguminous host plants, in Materialy Mezhdunarodnoi nauchnoprakticheskoi kon ferentsii “Biotekhnologiya: i kachestvo zhizni” (Proc. Int. SciPract. Conf. “Biotechnology and Quality of Life”), Moscow, 2014, vol. 2, p. 15.
Park, H.D., Lee, D.H., Hong, Y.H., Kang, D.H., Lee, Y.K., Song, J., Lee, S.Y., Kim, J.W., Ki, C.S., and Lee, Y.W., Three Korean patients with maple syrup urine disease: four novel mutations in the BCKDHA gene, Ann. Clin. Lab. Sci., 2011, vol. 41, no. 2, pp. 167–173.
Los’, D.A., Structure, regulation of expression, an functioning of fatty acid desaturases, Usp. Biol. Khim., 2001, vol. 41, pp. 163–198.
Aguilar, P.S. and de Mendoza, D., Control of fatty acid desaturation: a mechanism conserved from bacteria to humans, Mol. Microbiol., 2006, vol. 62, no. 6, pp. 1507–1514.
Pocard, J.A., Vincent, N., Boncompagni, E., Smith, L.T., Poggi, M.C., and Le Rudulier, D., Molecular character ization of the bet genes encoding glycine betaine synthesis in Sinorhizobium meliloti 102F34, Microbiology (UK), 1997, vol. 143, Pt 4, pp. 1369–1379.
Yurgel, S., Rice, J., Mulder, M., Kahn, M., Belova, V., and Roumiantseva, M., Truncated betB2144 plays a critical role in Sinorhizobium meliloti Rm2011 osmo protection and glycinebetaine catabolism, Eur. J. Soil Biol., 2013, vol. 54, pp. 48–55.
VelascoGarcia, R., GonzalezSegura, L., and Munoz-Clares, R.A., Steadystate kinetic mechanism of the NADP+-and NAD+-dependent reaction catalyzed by betaine aldehyde dehydrogenase from Pseudomonas aeruginosa, Biochem. J., 2000, vol. 352, pp. 675–683.
Cregut, M., Durand, M.J., and Thouand, G., The diversity and functions of choline sulphatases in micro organisms, Microb. Ecol., 2014, vol. 67, no. 2, pp. 350–357. doi: 10.1007/s0024801303287. Epub 2013.
MunozClares, R.A., DiazSanchez, A.G., Gonzalez-Segura, L., and Montiel, C., Kinetic and structural fea tures of betaine aldehyde dehydrogenases: mechanistic and regulatory implications, Arch. Biochem. Biophys., 2010, vol. 493, no. 1, pp. 71–81.
Smith, L.T., Pocard, J.A., Bernard, T., and Le Rudulier, D., Osmotic control of glycine betaine biosynthe sis and degradation in Rhizobium meliloti, J. Bacteriol., 1988, vol. 170, no. 7, pp. 3142–3149.
Canovas, D., Vargas, C., Kneip, S., Moron, M.J., Ventosa, A., Bremer, E., and Nieto, J.J., Characterization of the genes for the synthesis of the compatible solute glycine betaine in the moderately halophilic bacterium Halomonas elongate DSM 3043, Microbiology (UK), 2000, vol. 146, pp. 455–463.
Vargas, C., Argandona, M., ReinaBueno, M., RodriguezMoya, J., FernandezAunion, C., and Nieto, J.J., Unravelling the adaptation responses to osmotic and temperature stress in Chromohalobacter salexigens, a bac terium with broad salinity tolerance, Saline Systems, 2008, vol. 4, no. 14. doi:10.1186/17461448414
Mandon, K., Osteras, M., Boncompagni, E., Trinchant, J.C., Spennato, G., Poggi, M.C., and Le Rudulier, D., The Sinorhizobium meliloti glycine betaine biosynthetic genes (betlCBA) are induced by choline and highly expressed in bacteroids, Mol. Plant–Microbe Interact., 2003, vol. 16, no. 8, pp. 709–719.
Lamark, T., Rokenes, T.P., McDougall, J., and Strom, A.R., The complex bet promoters of Escheri chia coli: regulation by oxygen (ArcA), choline (BetI), and osmotic stress, J. Bacteriol., 1996, vol. 178, no. 6, pp. 1655–1662.
Ziegler, C., Bremer, E., and Kraemer, R., The BCCT family of carriers: from physiology to crystal structure, Mol. Microbiol., 2010, vol. 78, no. 1, pp. 13–34.
Wyn-Jones, R.G. and Storey, R., Betaines, in The Phys iology and Biochemistry of Drought Resistance in Plant, Paleg, I.G. and Aspinall, D., Eds., Sydney: Academic, 1981, pp. 117–204.
Osteras, M., Boncompagni, E., Vincent, N., Poggi, M.C., and Le Rudulier, D., Presence of a gene encoding choline sulfatase in Sinorhizobium meliloti bet operon: cholineO sulfate is metabolized into glycine betaine, Proc. Natl. Acad. Sci. U. S. A., 1998, vol. 95, pp. 11394–11399.
Selye, H.A., Syndrome produced by diverse nocuos agents, Nature, 1936, vol. 138, p. 32
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © M.L. Roumiantseva, V.S. Muntyan, 2015, published in Mikrobiologiya, 2015, Vol. 84, No. 3, pp. 263–280.
Rights and permissions
About this article
Cite this article
Roumiantseva, M.L., Muntyan, V.S. Root nodule bacteria Sinorhizobium meliloti: Tolerance to salinity and bacterial genetic determinants. Microbiology 84, 303–318 (2015). https://doi.org/10.1134/S0026261715030170
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026261715030170