Abstract
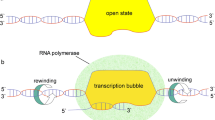
In this study we used the kink solutions of the nonlinear sine-Gordon equation to apply methods of mathematical modeling to investigate the dynamics of the transcription bubble in the pPF1 plasmid. Based on the calculated energy profile for the pPF1 plasmid and its modified versions, it was shown that the minimum potential energy of the kink formation or transcription bubble nucleation corresponds to the region between the genes of the Egfp and mCherry proteins. The insertion of homogeneous sequences into the region between Egfp and mCherry showed that the kink is more likely to be activated in polyT or polyC compared to polyA or polyG, which indicates the dependence of nucleation of the transcription bubble on the molecular weight of base pairs. For insertions into the region between Egfp and mCherry of small fragments of the native sequence of Escherichia coli, the model identifies the DNA strands with the highest probability of nucleation of the transcription bubble and, accordingly, determines the direction (towards the Egfp or mCherry gene) of transcription, indicating a link between the direction of transcription and the energy profile of the plasmid.













Similar content being viewed by others
REFERENCES
Map of plasmid pET-28b https://www.snapgene. com/resources/plasmid-files/?set=pet_and_duet_vectors_(novagen)&plasmid=pET-28b(%2B). Accessed November 1, 2020.
I. S. Masulis, Z. Sh. Babaeva, S. V. Chernyshov, et al., Sci. Rep. 5, 11449 (2015).
S. Borukhov and E. Nudler, Trends Microbiol. 16 (3), 126 (2008).
J. Y. Kang, T. V. Mishanina, R. Landick, et al., J. Mol. Biol. 431 (20), 4007 (2019).
M. T. Jr. Record, W. S. Reznikoff, M. L. Craig, et al., in Escherichia coli and Salmonella Cellular and Molecular Biology, Ed. by F.C. Neidhardt (ASM Press, Washington, DC, 1996), pp. 792–821.
H. Boyaci, J. Chen, R. Jansen, et al., Nature 565 (7739), 382 (2019).
J. Chen, C. Chiu, S. Gopalkrishnan, et al., Mol Cell. 78 (2), 275 (2020).
E. F. Ruff, M. T. Jr. Record, and I. Artsimovitch. Biomolecules 5 (2), 1035 (2015).
J. T. Winkelman, I. O. Vvedenskaya, Y. Zhang, et al., Science 351 (6277), 1090 (2016).
P. J. Caudrey, J. C. Eilbeck, and J. D. Gibbon, Nuovo Cimento B 25 (2), 497 (1975).
V. G. Ivancevic and T. T. Ivancevic, J. Geometry Symmetry Phys. 31, 1 (2013).
A. A. Grinevich, A. A. Ryasik, and L. V. Yakushevich, Chaos Soliton. Fract. 75, 62 (2015).
L. V. Yakushevich and L.A. Krasnobaeva, Math. Biol. Bioinform. 14 (1), 327 (2019).
K. S. Shavkunov, I. S. Masulis, M. N. Tutukina, et al., Nucleic Acids Res. 37 (15), 4919 (2009).
L. A. Krasnobaeva and L. V. Yakushevich, J. Bioinform. Comput. Biol. 13 (1), 1540002 (2015).
L. V. Yakushevich and L. A. Krasnobaeva, Int. J. Nonlinear Mech. 43, 1074 (2008).
L. V. Yakushevich, J. Biol. Phys. 43, 113 (2017).
W. Englander, N. R. Kallenbach, A. J. Heeger, et al., Proc. Natl. Acad. Sci. U. S. A. 77, 7222 (1980).
L. V. Yakushevich, L. A. Krasnobaeva, A. V. Sha-povalov, and N. R. Quintero, Biophysics (Moscow) 50 (3) 404 (2005).
L. V. Yakushevich and A. A Ryasik, Komp’yut. Issled. Moled. 4 (1), 209 (2012).
Y. Zhang, Y. Feng, S. Chatterjee, et al., Science 338 (6110), 1076 (2012).
E. Heyduk, K. Kuznedelov, K. Severinov, et al. J. Biol. Chem. 281 (18), 12362 (2006).
M. E. Karpen and P. L. deHaseth, Biomolecules 5 (2), 668 (2015).
N. S. Roy, S. Debnath, A. Chakraborty, et al., Phys. Chem. Chem. Phys. 20 (14), 9449 (2018).
F. Colizzi, C. Perez-Gonzalez, R. Fritzen, et al., Proc. Natl. Acad. Sci. U. S. A. 116 (45), 22471 (2019).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by E. Makeeva
Abbreviations: bp, base pairs.
Rights and permissions
About this article
Cite this article
Grinevich, A.A., Masulis, I.S. & Yakushevich, L.V. Mathematical Modeling of Transcription Bubble Behavior in the pPF1 Plasmid and its Modified Versions: The Link between the Plasmid Energy Profile and the Direction of Transcription. BIOPHYSICS 66, 209–217 (2021). https://doi.org/10.1134/S000635092102007X
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S000635092102007X