Abstract
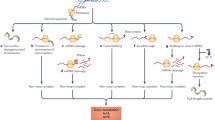
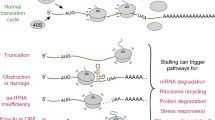
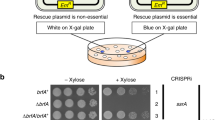
Translation of the genetic information into proteins, performed by the ribosome, is a key cellular process in all organisms. Translation usually proceeds smoothly, but, unfortunately, undesirable events can lead to stalling of translating ribosomes. To rescue these faulty arrested ribosomes, bacterial cells possess three well-characterized quality control systems, tmRNA, ArfA, and ArfB. Recently, an additional ribosome rescue mechanism has been discovered in Bacillus subtilis. In contrast to the “canonical” systems targeting the 70S bacterial ribosome, this latter mechanism operates by first splitting the ribosome into the small (30S) and large (50S) subunits to then clearing the resultant jammed large subunit from the incomplete nascent polypeptide. Here, I will discuss the recent microbiological, biochemical, and structural data regarding functioning of this novel rescue system.




Similar content being viewed by others
Abbreviations
- CAT tail:
-
Carboxy-terminal Alanine and Threonine tail
- RQC:
-
Ribosome-associated Quality Control
- tmRNA:
-
transfer-messenger RNA
References
Dai, X., Zhu, M., Warren, M., Balakrishnan, R., Patsalo, V., et al. (2016) Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth, Nat. Microbiol., 2, 16231.
Zhu, M., Dai, X., and Wang, Y.-P. (2016) Real time determination of bacterial in vivo ribosome translation elongation speed based on LacZα complementation system, Nucleic Acid Res., 44, e155.
Jha, S., and Komar, A. A. (2011) Birth, life and death of nascent polypeptide chains, Biotechnol. J., 6, 623-640.
Samatova, E., Daberger, J., Liutkute, M., and Rodnina, M. V. (2021) Translational control by ribosome pausing in bacteria: how a non-uniform pace of translation affects protein production and folding, Front. Microbiol., 11, 619430.
Doerfel, L. K., Wohlgemuth, I., Kothe, C., Peske, F., Urlaub, H., and Rodnina, M. V. (2013) EF-P is essential for rapid synthesis of proteins containing consecutive proline residues, Science, 339, 85-88.
Ude, S., Lassak, J., Starosta, A. L., Kraxenberger, T., Wilson, D. N., and Jung, K. (2013) Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches, Science, 339, 82-85.
Huter, P., Arenz, S., Bock, L. V., Graf, M., Frister, J. O., et al. (2017) Structural basis for polyproline-mediated ribosome stalling and rescue by the translation elongation factor EF-P, Mol. Cell, 68, 515-527.
Pech, M., Karim, Z., Yamamoto, H., Kitakawa, M., Qin, Y., and Nierhaus, K. H. (2011) Elongation factor 4 (EF4/LepA) accelerates protein synthesis at increased Mg2+ concentrations, Proc. Natl. Acad. Sci. USA, 108, 3199-3203.
Betney, R., de Silva, E., Krishnan, J., and Stansfield, I. (2010) Autoregulatory systems controlling translation factor expression: thermostat-like control of translational accuracy, RNA, 16, 655-663.
Shalgi, R., Hurt, J. A., Krykbaeva, I., Taipale, M., Lindquist, S., and Burge, C. B. (2013) Widespread regulation of translation by elongation pausing in heat shock, Mol. Cell, 49, 439-452.
Vazquez-Laslop, N., and Mankin, A. S. (2018) How macrolide antibiotics work, Trends Biochem. Sci., 43, 668-684.
Muller, C., Crowe-McAuliffe, C., and Wilson, D. N. (2021) Ribosome rescue pathways in bacteria, Front. Microbiol., 12, 652980.
Buskirk, A. R., and Green, R. (2017) Ribosome pausing, arrest and rescue in bacteria and eukaryotes, Philos. Trans. R. Soc. Lond. B. Biol. Sci., 372, 20160183.
Keiler, K. C. (2015) Mechanisms of ribosome rescue in bacteria, Nat. Rev. Microbiol., 13, 285-297.
Keiler, K. C., Waller, P. R., and Sauer, R. T. (1996) Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA, Science, 271, 990-993.
Karzai, A. W., Susskind, M. M., and Sauer, R. T. (1999) SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA), EMBO J., 18, 3793-3799.
Himeno, H., Nameki, N., Kurita, D., Muto, A., and Abo, T. (2015) Ribosome rescue systems in bacteria, Biochimie, 114, 102-112.
Chadani, Y., Ito, K., Kutsukake, K., and Abo, T. (2012) ArfA recruits release factor 2 to rescue stalled ribosomes by peptidyl-tRNA hydrolysis in Escherichia coli, Mol. Microbiol., 86, 37-50.
Shimizu, Y. (2012) ArfA recruits RF2 into stalled ribosomes, J. Mol. Biol., 423, 624-631.
Goralski, T. D. P., Kirimanjeswara, G. S., and Keiler, K. C. (2018) A new mechanism for ribosome rescue can recruit RF1 or RF2 to nonstop ribosomes, mBio, 9, e02436-18.
Handa, Y., Inaho, N., and Nameki, N. (2011) YaeJ is a novel ribosome-associated protein in Escherichia coli that can hydrolyze peptidyl-tRNA on stalled ribosomes, Nucleic Acids Res., 39, 1739-1748.
Gagnon, M. G., Seetharaman, S. V., Bulkley, D., and Steitz, T. A. (2012) Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome, Science, 335, 1370-1372.
Shoemaker, C. J., and Green, R. (2012) Translation drives mRNA quality control, Nat. Struct. Mol. Biol., 19, 594-601.
Pisarev, A. V., Skabkin, M. A., Pisareva, V. P., Skabkina, O. V., Rakotondrafara, A. M., et al. (2010) The role of ABCE1 in eukaryotic post-termination ribosomal recycling, Mol. Cell, 37, 196-210.
Shoemaker, C. J., Eyler, D. E., and Green, R. (2010) Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay, Science, 330, 369-372.
Joazeiro, C. A. P. (2019) Mechanisms and functions of ribosome-associated protein quality control, Nat. Rev. Mol. Cell Biol., 20, 368-383.
Shen, P. S., Park, J., Qin, Y., Li, X., Parsawar, K., et al. (2015) Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains, Science, 347, 75-78.
Verma, R., Reichermeier, K. M., Burroughs, A. M., Oania, R. S., Reitsma, J. M., et al. (2018) Vms1/ANKZF1 peptidyl-tRNA hydrolases releases nascent chains from stalled ribosomes, Nature, 557, 446-451.
Yip, M. C. J., Keszei, A. F. A., Feng, Q., Chu, V., McKenna, M. J., and Shao, S. (2019) Mechanism for recycling tRNAs on stalled ribosomes, Nat. Struct. Mol. Biol., 26, 343-349.
Yip, M. C. J., Savickas, S., Gygi, S. P., and Shao, S. (2020) ELAC1 repairs tRNA cleaved during ribosome-associated quality control, Cell Rep., 30, 2106-2114.
Chadani, Y., Ono, K., Kutsukake, K., and Abo, T. (2011) Escherichia coli YaeJ protein mediates a novel ribosome-rescue pathway distinct from SsrA- and ArfA-mediated pathways, Mol. Microbiol., 80, 772-785.
Keiler, K. C., and Feaga, H. A. (2014) Resolving nonstop translation complexes is a matter of life or death, J. Bacteriol., 196, 2123-2130.
Burroughs, A. M., and Aravind, L. (2014) Analysis of two domains with novel RNA-processing activities throws light on the complex evolution of ribosomal RNA biogenesis, Front. Genet., 5, 424.
Henderson, B., Nair, S., Pallas, J., and Williams, M. A. (2011) Fibronectin: a multidomain host adhesin targeted by bacterial fibronectin-binding proteins, FEMS Microbiol. Rev., 35, 147-200.
Lytvynenko, I., Paternoga, H., Thrun, A., Balke, A., Muller, T. A., et al. (2019) Alanine tails signal proteolysis in bacterial ribosome-associated quality control, Cell, 178, 76-90.
Filbeck, S., Cerullo, F., Paternoga, H., Tsaprailis, G., Joazeiro, C. A. P., and Pfeffer, S. (2021) Mimicry of canonical translation elongation underlies alanine tail synthesis in RQC, Mol. Cell, 81, 104-114.
Crowe-McAuliffe, C., Takada, H., Murina, V., Polte, C., Kasvandik, S., et al. (2021) Structural basis for bacterial ribosome-associated quality control by RqcH and RqcP, Mol. Cell, 81, 115-126.
Chadani, Y., Niwa, T., Izumi, T., Sugata, N., Nagao, A., et al. (2017) Intrinsic ribosome destabilization underlies translation and provides an organism with a strategy of environmental sensing, Mol. Cell, 68, 528-539.
Chadani, Y., Sugata, N., Niwa, T., Ito, Y., Iwasaki, S., and Taguchi, H. (2021) Nascent polypeptide within the exit tunnel ensures continuous translation elongation by stabilizing the translating ribosome, bioRxiv, https://doi.org/10.1101/2021.02.02.429294.
Peske, F., Rodnina, M. V., and Wintermeyer, W. (2005) Sequence of steps in ribosome recycling as defined by kinetic analysis, Mol. Cell, 18, 403-412.
Zhang, Y., Mandava, C. S., Cao, W., Li, X., Zhang, D., et al. (2015) HflX is a ribosome-splitting factor rescuing stalled ribosomes under stress conditions, Nat. Struct. Mol. Biol., 22, 906-913.
Tomar, S. K., Kumar, P., and Prakash, B. (2011) Deciphering the catalytic machinery in a universally conserved ribosome binding ATPase YchF, Biochem. Biophys. Res. Commun., 408, 459-464.
Feng, B., Mandava, C. S., Guo, Q., Wang, J., Cao, W., et al. (2014) Structural and functional insights into the mode of action of a universally conserved Obg GTPase, PLos Biol., 12, e1001866.
Korber, P., Stahl, J. M., Nierhaus, K. H., and Bardwell, J. C. (2000) Hsp15: a ribosome-associated heat shock protein, EMBO J., 19, 741-748.
Jiang, L., Schaffitzel, C., Bingel-Erlenmeyer, R., Ban, N., Korber, P., et al. (2009) Recycling of aborted ribosomal 50S subunit-nascent chain-tRNA complexes by the heat shock protein Hsp15, J. Mol. Biol., 386, 1357-1367.
Starosta, A. L., Lassak, J., Jung, K., and Wilson, D. N. (2014) The bacterial translation stress response, FEMS Microbiol. Rev., 38, 1172-1201.
Das, G., and Varshney, U. (2006) Peptidyl-tRNA hydrolase and its critical role in protein biosynthesis, Microbiology, 152, 2191-2195.
Sharma, S., Kaushik, S., Sinha, M., Kushwaha, G. S., Singh, A., et al. (2014) Structural and functional insights into peptidyl-tRNA hydrolase, Biochim. Biophys. Acta, 1844, 1279-1288.
Burroughs, A. M., and Aravind, L. (2019) The origin and evolution of release factors: implications for translation termination, ribosome rescue, and quality control pathways, Int. J. Mol. Sci., 20, 1981.
Lewis, K. (2020) The science of antibiotic discovery, Cell, 181, 29-45.
Holmes, A. R., McNab, R., Millsap, K. W., Rohde, M., Hammerschmidt, S., et al. (2001) The pavA gene of Streptococcus pneumoniae encodes a fibronectin-binding protein that is essential for virulence, Mol. Microbiol., 41, 1395-1408.
Pracht, D., Elm, C., Gerber, J., Bergmann, S., Rohde, M., et al. (2005) PavA of Streptococcus pneumoniae modulates adherence, invasion, and meningeal inflammation, Infect. Immun., 73, 2680-2689.
Singh, K. V., La Rossa, S. L., Somarajan, S. R., Roh, J. H., and Murray, B. E. (2015) The fibronectin-binding protein EfbA contributes to pathogenesis and protects against infective endocarditis caused by Enterococcus faecalis, Infect. Immun., 83, 4487-4494.
Dramsi, S., Bourdichon, F., Cabanes, D., Lecuit, M., Fsihi, H., and Cossart, P. (2004) FbpA, a novel multifunctional Listeria monocytogenes virulence factor, Mol. Microbiol., 53, 639-649.
Ramadoss, N. S., Alumasa, J. N., Cheng, L., Wang, Y., Li, S., et al. (2013) Small molecule inhibitors of trans-translation have broad-spectrum antibiotic activity, Proc. Natl. Acad. Sci. USA, 110, 10282-10287.
Aron, Z. D., Mehrani, A., Hoffer, E. D., Connolly, K. L., Srinivas, P., et al. (2021) trans-Translation inhibitors bind to a novel site on the ribosome and clear Neisseria gonorrhoeae in vivo, Nat. Commun., 12, 1799.
Acknowledgments
I want to express my deepest gratitude to my teacher Dr. Alexander Spirin and members of the Institute of Protein Research in Pushchino, where I gained priceless experience in the fields of protein biosynthesis and ribosomology. I am grateful to Alexander Richardson, Dr. Yury Polikanov, Dr. Nora Vázquez-Laslop, and Dr. Alexander Mankin for critical reading and fruitful discussion of the review.
Funding
This work was supported by the NIH grant R21-AI137584.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author declares no conflicts of interest in financial or any other sphere. This article does not contain any studies involving human participants or animals performed by the author.
Rights and permissions
About this article
Cite this article
Svetlov, M.S. Ribosome-Associated Quality Control in Bacteria. Biochemistry Moscow 86, 942–951 (2021). https://doi.org/10.1134/S0006297921080058
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297921080058