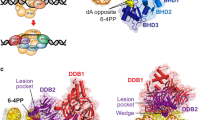
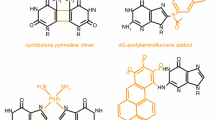
Abstract
Nucleotide excision repair (NER) is one of the major DNA repair pathways aimed at maintaining genome stability. Correction of DNA damage by the NER system is a multistage process that proceeds with the formation of multiple DNA-protein and protein-protein intermediate complexes and requires precise coordination and regulation. NER proteins undergo post-translational modifications, such as ubiquitination, sumoylation, phosphorylation, acetylation, and poly(ADP-ribosyl)ation. These modifications affect the interaction of NER factors with DNA and other proteins and thus regulate either their recruitment into the complexes or dissociation from these complexes at certain stages of DNA repair, as well as modulate the functional activity of NER proteins and control the process of DNA repair in general. Here, we review the data on the post-translational modifications of NER factors and their effects on DNA repair. Protein poly(ADP-ribosyl)ation catalyzed by poly(ADP-ribose) polymerase 1 and its impact on NER are discussed in detail, since such analysis has not been done before.
Similar content being viewed by others
Abbreviations
- BPDE:
-
benz[a]pyrene diol epoxide
- CPD:
-
cyclobutane pyrimidine dimer
- GG-NER:
-
global genome nucleotide excision repair
- NER:
-
nucleotide excision repair
- PAR:
-
poly(ADP-ribose)
- PARP:
-
poly(ADP-ribose) polymerase
- RNAP:
-
RNA polymerase
- RPA:
-
replication protein A
- SUMO:
-
small ubiquitin-like modifier
- TC-NER:
-
transcription-coupled NER
- TFIIH:
-
transcription factor IIH
- XP:
-
xeroderma pigmentosum
- XPC and XPA:
-
xeroderma pigmentosum factors C and A, respectively
References
Friedberg, E. C. (2003) DNA damage and repair, Nature, 421, 436–440, doi: https://doi.org/10.1038/nature01408.
Gillet, L. C., and Scharer, O. D. (2006) Molecular mechanisms of mammalian global genome nucleotide excision repair, Chem. Rev., 106, 253–276, doi: https://doi.org/10.1021/cr040483f.
Tornaletti, S., and Hanawalt, P. C. (1999) Effect of DNA lesions on transcription elongation, Biochimie, 81, 139–146.
Fousteri, M., and Mullenders, L. H. (2008) Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects, Cell Res., 18, 73–84, doi: https://doi.org/10.1038/cr.2008.6.
Sugasawa, K., Shimizu, Y., Iwai, S., and Hanaoka, F. (2002) A molecular mechanism for DNA damage recognition by the xeroderma pigmentosum group C protein complex, DNA Repair (Amst.), 1, 95–107.
Maillard, O., Camenisch, U., Clement, F. C., Blagoev, K. B., and Naegeli, H. (2007) DNA repair triggered by sensors of helical dynamics, Trends Biochem. Sci., 32, 494–499, doi: https://doi.org/10.1016/j.tibs.2007.08.008.
Min, J. H., and Pavletich, N. P. (2007) Recognition of DNA damage by the Rad4 nucleotide excision repair protein, Nature, 449, 570–575, doi: https://doi.org/10.1038/nature06155.
Maltseva, E. A., Rechkunova, N. I., Petruseva, I. O., Vermeulen, W., Scharer, O. D., and Lavrik, O. I. (2008) Crosslinking of nucleotide excision repair proteins with DNA containing photoreactive damages, Bioorg. Chem., 36, 77–84, doi: https://doi.org/10.1016/j.bioorg.2007.11.004.
Rechkunova, N. I., and Lavrik, O. I. (2010) Nucleotide excision repair in higher eukaryotes: mechanism of primary damage recognition in global genome repair, Subcell. Biochem., 50, 251–277, doi: https://doi.org/10.1007/978-90-481-3471-7_13.
Krasikova, Y. S., Rechkunova, N. I., Maltseva, E. A., Pestryakov, P. E., Petruseva, I. O., Sugasawa, K., Chen, X., Min, J. H., and Lavrik, O. I. (2013) Comparative analysis of interaction of human and yeast DNA damage recognition complexes with damaged DNA in nucleotide excision repair, J. Biol. Chem., 288, 10936–10947, doi: https://doi.org/10.1074/jbc.M112.444026.
Fitch, M. E., Nakajima, S., Yasui, A., and Ford, J. M. (2003) In vivo recruitment of XPC to UV-induced cyclobutane pyrimidine dimers by the DDB2 gene product, J. Biol. Chem., 278, 46906–46910, doi: https://doi.org/10.1074/jbc.M307254200 https://doi.org/10.1074/jbc.M307254200.
Moser, J., Volker, M., Kool, H., Alekseev, S., Vrieling, H., Yasui, A., van Zeeland, A. A., and Mullenders, L. H. (2005) The UV-damaged DNA binding protein mediates efficient targeting of the nucleotide excision repair complex to UV-induced photo lesions, DNA Repair (Amst.), 4, 571–582, doi: https://doi.org/10.1016/j.dnarep.2005.01.001.
Houten, B. V., Kuper, J., and Kisker, C. (2016) Role of XPD in cellular functions: to TFIIH and beyond, DNA Repair (Amst.), 44, 136–142, doi: https://doi.org/10.1016/j.dnarep.2016.05.019.
Evans, E., Moggs, J. G., Hwang, J. R., Egly, J. M., and Wood, R. D. (1997) Mechanism of open complex and dual incision formation by human nucleotide excision repair factors, EMBO J., 16, 6559–6573, doi: https://doi.org/10.1093/emboj/16.21.6559.
Staresincic, L., Fagbemi, A. F., Enzlin, J. H., Gourdin, A. M., Wijgers, N., Dunand-Sauthier, I., Giglia-Mari, G., Clarkson, S. G., Vermeulen, W., and Scharer, O. D. (2009) Coordination of dual incision and repair synthesis in human nucleotide excision repair, EMBO J., 28, 1111–1120, doi: https://doi.org/10.1038/emboj.2009.49.
Krasikova, Y. S., Rechkunova, N. I., Maltseva, E. A., Petruseva, I. O., and Lavrik, O. I. (2010) Localization of xeroderma pigmentosum group A protein and replication protein A on damaged DNA in nucleotide excision repair, Nucleic Acids Res., 38, 8083–8094, doi: https://doi.org/10.1093/nar/gkq649.
Kemp, M. G., Gaddameedhi, S., Choi, J. H., Hu, J., and Sancar, A. (2014) DNA repair synthesis and ligation affect the processing of excised oligonucleotides generated by human nucleotide excision repair, J. Biol. Chem., 289, 26574–26583, doi: https://doi.org/10.1074/jbc.M114.597088.
Grabbe, C., Husnjak, K., and Dikic, I. (2011) The spatial and temporal organization of ubiquitin networks, Nat. Rev. Mol. Cell. Biol., 12, 295–307, doi: https://doi.org/10.1038/nrm3099.
Husnjak, K., and Dikic, I. (2012) Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions, Annu. Rev. Biochem., 81, 291–322, doi: https://doi.org/10.1146/annurevbiochem-051810-094654.
Komander, D., and Rape, M. (2012) The ubiquitin code, Annu. Rev. Biochem., 81, 203–229, doi: https://doi.org/10.1146/annurev-biochem-060310-170328.
Van Cuijk, L., Vermeulen, W., and Marteijn, J. A. (2014) Ubiquitin at work: the ubiquitous regulation of the damage recognition step of NER, Exp. Cell. Res., 329, 101–109, doi: https://doi.org/10.1016/j.yexcr.2014.07.018.
Ruthemann, P., Balbo Pogliano, C., and Naegeli, H. (2016) Global-genome nucleotide excision repair controlled by ubiquitin/sumo modifiers, Front. Genet., 7, 68, doi: https://doi.org/10.3389/fgene.2016.00068.
Chitale, S., and Richly, H. (2017) Timing of DNA lesion recognition: ubiquitin signaling in the NER pathway, Cell Cycle, 16, 163–171, doi: https://doi.org/10.1080/15384101.2016.1261227.
Sugasawa, K., Okuda, Y., Saijo, M., Nishi, R., Matsuda, N., Chu, G., Mori, T., Iwai, S., Tanaka, K., Tanaka, K., and Hanaoka, F. (2005) UV-induced ubiquitination of XPC protein mediated by UV-DDB-ubiquitin ligase complex, Cell, 121, 387–400, doi: https://doi.org/10.1016/j.cell.2005.02.035.
Fischer, E. S., Scrima, A., Bohm, K., Matsumoto, S., Lingaraju, G. M., Faty, M., Yasuda, T., Cavadini, S., Wakasugi, M., Hanaoka, F., Iwai, S., Gut, H., Sugasawa, K., and Thoma, N. H. (2011) The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation, Cell, 147, 1024–1039, doi: https://doi.org/10.1016/j.cell.2011.10.035.
Kapetanaki, M. G., Guerrero-Santoro, J., Bisi, D. C., Hsieh, C. L., Rapic-Otrin, V., and Levine, A. S. (2006) The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites, Proc. Natl. Acad. Sci. USA, 103, 2588–2593, doi: https://doi.org/10.1073/pnas.0511160103.
Wang, H., Zhai, L., Xu, J., Joo, H. Y., Jackson, S., Erdjument-Bromage, H., Tempst, P., Xiong, Y., and Zhang, Y. (2006) Histone H3 and H4 ubiquitination by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage, Mol. Cell, 22, 383–394, doi: https://doi.org/10.1016/j.molcel.2006.03.035.
Groisman, R., Polanowska, J., Kuraoka, I., Sawada, J., Saijo, M., Drapkin, R., Kisselev, A. F., Tanaka, K., and Nakatani, Y. (2003) The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage, Cell, 113, 357–367.
Matsumoto, S., Fischer, E. S., Yasuda, T., Dohmae, N., Iwai, S., Mori, T., Nishi, R., Yoshino, K., Sakai, W., Hanaoka, F., Thoma, N. H., and Sugasawa, K. (2015) Functional regulation of the DNA damage-recognition factor DDB2 by ubiquitination and interaction with xeroderma pigmentosum group C protein, Nucleic Acids Res., 43, 1700–1713, doi: https://doi.org/10.1093/nar/gkv038.
Puumalainen, M. R., Lessel, D., Ruthemann, P., Kaczmarek, N., Bachmann, K., Ramadan, K., and Naegeli, H. (2014) Chromatin retention of DNA damage sensors DDB2 and XPC through loss of p97 segregase causes genotoxicity, Nat. Commun., 5, 3695, doi: https://doi.org/10.1038/ncomms4695.
He, J., Zhu, Q., Wani, G., Sharma, N., Han, C., Qian, J., Pentz, K., Wang, Q. E., and Wani, A. A. (2014) Ubiquitin-specific protease 7 regulates nucleotide excision repair through deubiquitinating XPC protein and preventing XPC protein from undergoing ultraviolet light-induced and VCP/p97 protein-regulated proteolysis, J. Biol. Chem., 289, 27278–27289, doi: https://doi.org/10.1074/jbc.M114.589812.
Guerrero-Santoro, J., Kapetanaki, M. G., Hsieh, C. L., Gorbachinsky, I., Levine, A. S., and Rapic-Otrin, V. (2008) The cullin 4B-based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A, Cancer Res., 68, 5014–5022, doi: https://doi.org/10.1158/0008-5472.CAN-07-6162.
Wang, Q. E., Zhu, Q., Wani, G., El-Mahdy, M. A., Li, J., and Wani, A. A. (2005) DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation, Nucleic Acids Res., 33, 4023–4034, doi: https://doi.org/10.1093/nar/gki684.
Van Cuijk, L., van Belle, G. J., Turkyilmaz, Y., Poulsen, S. L., Janssens, R. C., Theil, A. F., Sabatella, M., Lans, H., Mailand, N., Houtsmuller, A. B., Vermeulen, W., and Marteijn, J. A. (2015) SUMO and ubiquitin-dependent XPC exchange drives nucleotide excision repair, Nat. Commun., 6, 7499, doi: https://doi.org/10.1038/ncomms8499.
Han, C., Zhao, R., Kroger, J., He, J., Wani, G., Wang, Q. E., and Wani, A. A. (2017) UV irradiation-induced SUMOylation of DDB2 regulates nucleotide excision repair, Carcinogenesis, 38, 976–985, doi: https://doi.org/10.1093/carcin/bgx076.
Poulsen, S. L., Hansen, R. K., Wagner, S. A., van Cuijk, L., van Belle, G. J., Streicher, W., Wikstrom, M., Choudhary, C., Houtsmuller, A. B., Marteijn, J. A., Bekker-Jensen, S., and Mailand, N. (2013) RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response, J. Cell. Biol., 201, 797–807, doi: https://doi.org/10.1083/jcb.201212075.
Akita, M., Tak, Y. S., Shimura, T., Matsumoto, S., Okuda-Shimizu, Y., Shimizu, Y., Nishi, R., Saitoh, H., Iwai, S., Mori, T., Ikura, T., Sakai, W., Hanaoka, F., and Sugasawa, K. (2015) SUMOylation of xeroderma pigmentosum group C protein regulates DNA damage recognition during nucleotide excision repair, Sci. Rep., 5, 10984, doi: https://doi.org/10.1038/srep10984.
Klein, U. R., and Nigg, E. A. (2009) SUMO-dependent regulation of centrin-2, J. Cell. Sci., 122, 3312–3321, doi: https://doi.org/10.1242/jcs.050245.
Wilson, M. D., Harreman, M., and Svejstrup, J. Q. (2013) Ubiquitination and degradation of elongating RNA polymerase II: the last resort, Biochim. Biophys. Acta, 1829, 151–157, doi: https://doi.org/10.1016/j.bbagrm.2012.08.002.
Kang, T. H., Lindsey-Boltz, L. A., Reardon, J. T., and Sancar, A. (2010) Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase, Proc. Natl. Acad. Sci. USA, 107, 4890–4895, doi: https://doi.org/10.1073/pnas.0915085107.
Yates, M., and Marechal, A. (2018) Ubiquitination at the fork: making and breaking chains to complete DNA replication, Int. J. Mol. Sci., 19, E2909, doi: https://doi.org/10.3390/ijms19102909.
Dou, H., Huang, C., Singh, M., Carpenter, P. B., and Yeh, E. T. (2010) Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex, Mol. Cell, 39, 333–345, doi: https://doi.org/10.1016/j.molcel.2010.07.021.
Perez-Oliva, A. B., Lachaud, C., Szyniarowski, P., Munoz, I., Macartney, T., Hickson, I., Rouse, J., and Alessi, D. R. (2015) USP45 deubiquitinase controls ERCC1-XPF endonuclease-mediated DNA damage responses, EMBO J., 34, 326–343, doi: https://doi.org/10.15252/embj.201489184.
Ame, J. C., Spenlehauer, C., and de Murcia, G. (2004) The PARP superfamily, Bioessays, 26, 882–893, doi: https://doi.org/10.1002/bies.20085.
Schreiber, V., Dantzer, F., Ame, J. C., and de Murcia, G. (2006) Poly(ADP-ribose): novel functions for an old molecule, Nat. Rev. Mol. Cell. Biol., 7, 517–528, doi: https://doi.org/10.1038/nrm1963.
Shieh, W. M., Ame, J. C., Wilson, M. V., Wang, Z. Q., Koh, D. W., Jacobson, M. K., and Jacobson, E. L. (1998) Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J. Biol. Chem., 273, 30069–30072.
Virag, L., and Szabo, C. (2002) The therapeutic potential of poly(ADP-ribose) polymerase inhibitors, Pharmacol. Rev., 54, 375–429.
Burkle, A., and Virag, L. (2013) Poly(ADP-ribose): PARadigms and PARadoxes, Mol. Aspects Med., 34, 1046–1065, doi: https://doi.org/10.1016/j.mam.2012.12.010.
Kraus, W. L., and Hottiger, M. O. (2013) PARP-1 and gene regulation: progress and puzzles, Mol. Aspects Med., 34, 1109–1123, doi: https://doi.org/10.1016/j.mam.2013.01.005.
Bock, F. J., Todorova, T. T., and Chang, P. (2015) RNA regulation by poly(ADP-ribose) polymerases, Mol. Cell, 58, 959–969, doi: https://doi.org/10.1016/j.molcel.2015.01.037.
Liu, C., Vyas, A., Kassab, M. A., Singh, A. K., and Yu, X. (2017) The role of poly ADP-ribosylation in the first wave of DNA damage response, Nucleic Acids Res., 45, 8129–8141, doi: https://doi.org/10.1093/nar/gkx565.
Khodyreva, S. N., and Lavrik, O. I. (2016) Poly(ADP-ribose) polymerase 1 as a key regulator of DNA repair, Mol. Biol. (Moscow), 50, 580–595, doi: https://doi.org/10.7868/S0026898416040030.
Sukhanova, M. V., Khodyreva, S. N., Lebedeva, N. A., Prasad, R., Wilson, S. H., and Lavrik, O. I. (2005) Human base excision repair enzymes apurinic/apyrimidinic endonuclease 1 (APE1), DNA polymerase beta and poly(ADP-ribose) polymerase 1: interplay between strand-displacement DNA synthesis and proofreading exonuclease activity, Nucleic Acids Res., 33, 1222–1229, doi: https://doi.org/10.1093/nar/gki266.
Sukhanova, M. V., Khodyreva, S. N., and Lavrik, O. I. (2004) Poly(ADP-ribose) polymerase-1 inhibits strand-displacement synthesis of DNA catalyzed by DNA polymerase beta, Biochemistry (Moscow), 69, 558–568.
Berger, N. A., Sikorski, G. W., Petzold, S. J., and Kurohara, K. K. (1980) Defective poly(adenosine diphosphoribose) synthesis in xeroderma pigmentosum, Biochemistry, 19, 289–293.
McCurry, L. S., and Jacobson, M. K. (1981) Poly(ADP-ribose) synthesis following DNA damage in cells heterozygous or homozygous for the xeroderma pigmentosum genotype, J. Biol. Chem., 256, 551–553.
Jacobson, E. L., Antol, K. M., Juarez-Salinas, H., and Jacobson, M. K. (1983) Poly(ADP-ribose) metabolism in ultraviolet irradiated human fibroblasts, J. Biol. Chem., 258, 103–107.
Yoon, Y. S., Kim, J. W., Kang, K. W., Kim, Y. S., Choi, K. H., and Joe, C. O. (1996) Poly(ADP-ribosyl)ation of histone H1 correlates with internucleosomal DNA fragmentation during apoptosis, J. Biol. Chem., 271, 9129–9134.
Chang, H., Sander, C. S., Muller, C. S., Elsner, P., and Thiele, J. J. (2002) Detection of poly(ADP-ribose) by immunocytochemistry: a sensitive new method for the early identification of UVB- and H2O2-induced apoptosis in keratinocytes, Biol. Chem., 383, 703–708, doi: https://doi.org/10.1515/BC.2002.072.
Pleschke, J. M., Kleczkowska, H. E., Strohm, M., and Althaus, F. R. (2000) Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins, J. Biol. Chem., 275, 40974–40980, doi: https://doi.org/10.1074/jbc.M006520200.
Fahrer, J., Kranaster, R., Altmeyer, M., Marx, A., and Burkle, A. (2007) Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length, Nucleic Acids Res., 35, e143, doi: https://doi.org/10.1093/nar/gkm944.
Gagne, J. P., Isabell, M., Lo, K. S., Bourassa, S., Hendzel, M. J., Dawson, V. L., Dawson, T. M., and Poirier, G. G. (2008) Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes, Nucleic Acids Res., 36, 6959–6976, doi: https://doi.org/10.1093/nar/gkn771.
Jungmichel, S., Rosenthal, F., Altmeyer, M., Lukas, J., Hottiger, M. O., and Nielsen, M. L. (2013) Proteome-wide identification of poly(ADP-ribosyl)ation targets in different genotoxic stress responses, Mol. Cell., 52, 272–285, doi: https://doi.org/10.1016/j.molcel.2013.08.026.
Flohr, C., Burkle, A., Radicella, J. P., and Epe, B. (2003) Poly(ADP-ribosyl)ation accelerates DNA repair in a pathway dependent on Cockayne syndrome B protein, Nucleic Acids Res., 31, 5332–5337.
Vodenicharov, M. D., Ghodgaonkar, M. M., Halappanavar, S. S., Shah, R. G., and Shah, G. M. (2005) Mechanism of early biphasic activation of poly(ADP-ribose) polymerase-1 in response to ultraviolet B irradiation, J. Cell. Sci., 118, 589–599, doi: https://doi.org/10.1242/jcs.01636.
Purohit, N. K., Robu, M., Shah, R. G., Geacintov, N. E., and Shah, G. M. (2016) Characterization of the interactions of PARP-1 with UV-damaged DNA in vivo and in vitro, Sci. Rep., 6, 19020, doi: https://doi.org/10.1038/srep19020.
Lin, T., and Yang, M. S. (2008) Benzo[a]pyrene-induced necrosis in the HepG(2) cells via PARP-1 activation and NAD+ depletion, Toxicology, 245, 147–153, doi: https://doi.org/10.1016/j.tox.2007.12.020.
Tao, G. H., Yang, L. Q., Gong, C. M., Huang, H. Y., Liu, J. D., Liu, J. J., Yuan, J. H., Chen, W., and Zhuang, Z. X. (2009) Effect of PARP-1 deficiency on DNA damage and repair in human bronchial epithelial cells exposed to benzo(a)pyrene, Mol. Biol. Rep., 36, 2413–2422, doi: https://doi.org/10.1007/s11033-009-9472-z.
Fischer, J. M. F., Zubel, T., Jander, K., Fix, J., Trussina, I. R. E. A., Gebhard, D., Bergemann, J., Burkle, A., and Mangerich, A. (2018) PARP1 protects from benzo[a]pyrene diol epoxide-induced replication stress and mutagenicity, Arch. Toxicol., 92, 1323–1340, doi: https://doi.org/10.1007/s00204-017-2115-6.
Pines, A., Vrouwe, M. G., Marteijn, J. A., Typas, D., Luijsterburg, M. S., Cansoy, M., Hensbergen, P., Deelder, A., de Groot, A., Matsumoto, S., Sugasawa, K., Thoma, N., Vermeulen, W., Vrieling, H., and Mullenders, L. (2012) PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1, J. Cell. Biol., 199, 235–249, doi: https://doi.org/10.1083/jcb.201112132.
Luijsterburg, M. S., Lindh, M., Acs, K., Vrouwe, M. G., Pines, A., van Attikum, H., Mullenders, L. H., and Dantuma, N. P. (2012) DDB2 promotes chromatin decondensation at UV-induced DNA damage, J. Cell Biol., 197, 267–281, doi: https://doi.org/10.1083/jcb.201106074.
Robu, M., Shah, R. G., Petitclerc, N., Brind'Amour, J., Kandan-Kulangara, F., and Shah, G. M. (2013) Role of poly(ADP-ribose) polymerase-1 in the removal of UV-induced DNA lesions by nucleotide excision repair, Proc. Natl. Acad. Sci. USA, 110, 1658–1663, doi: https://doi.org/10.1073/pnas.1209507110.
Robu, M., Shah, R. G., Purohit, N. K., Zhou, P., Naegeli, H., and Shah, G. M. (2017) Poly(ADP-ribose) polymerase 1 escorts XPC to UV-induced DNA lesions during nucleotide excision repair, Proc. Natl. Acad. Sci. USA, 114, 6847–6856, doi: https://doi.org/10.1073/pnas.1706981114.
Maltseva, E. A., Rechkunova, N. I., Sukhanova, M. V., and Lavrik, O. I. (2015) Poly(ADP-ribose) polymerase 1 modulates interaction of the nucleotide excision repair factor XPC-RAD23B with DNA via poly(ADP-ribosyl)ation, J. Biol. Chem., 290, 21811–21820, doi: https://doi.org/10.1074/jbc.M115.646638.
King, B. S., Cooper, K. L., Liu, K. J., and Hudson, L. G. (2012) Poly(ADP-ribose) contributes to an association between poly(ADP-ribose) polymerase-1 and xeroderma pigmentosum complementation group A in nucleotide excision repair, J. Biol. Chem., 287, 39824–39833, doi: https://doi.org/10.1074/jbc.M112.393504.
Fischer, J. M., Popp, O., Gebhard, D., Veith, S., Fischbach, A., Beneke, S., Leitenstorfer, A., Bergemann, J., Scheffner, M., Ferrando-May, E., Mangerich, A., and Burkle, A. (2014) Poly(ADP-ribose)-mediated interplay of XPA and PARP1 leads to reciprocal regulation of protein function, FEBS J., 281, 3625–3641, doi: https://doi.org/10.1111/febs.12885.
Eki, T., and Hurwitz, J. (1991) Influence of poly(ADP-ribose) polymerase on the enzymatic synthesis of SV40 DNA, J. Biol. Chem., 266, 3087–3100.
Gagne, J. P., Pic, E., Isabelle, M., Krietsch, J., Ethier, C., Paquet, E., Kelly, I., Boutin, M., Moon, K. M., Foster, L. J., and Poirier, G. G. (2012) Quantitative proteomics profiling of the poly(ADP-ribose)-related response to genotoxic stress, Nucleic Acids Res., 40, 7788–7805, doi: https://doi.org/10.1093/nar/gks486.
Illuzzi, G., Fouquerel, E., Ame, J. C., Noll, A., Rehmet, K., Nasheuer, H. P., Dantzer, F., and Schreiber, V. (2014) PARG is dispensable for recovery from transient replicative stress but required to prevent detrimental accumulation of poly(ADP-ribose) upon prolonged replicative stress, Nucleic Acids Res., 42, 7776–7792, doi: https://doi.org/10.1093/nar/gku505.
Maltseva, E. A., Krasikova, Y. S., Sukhanova, M. V., Rechkunova, N. I., and Lavrik, O. I. (2018) Replication protein A as a modulator of the poly(ADP-ribose)polymerase 1 activity, DNA Repair (Amst.), 72, 28–38, doi: https://doi.org/10.1016/j.dnarep.2018.09.010.
Thorslund, T., von Kobbe, C., Harrigan, J. A., Indig, F. E., Christiansen, M., Stevnsner, T., and Bohr, V. A. (2005) Cooperation of the Cockayne syndrome group B protein and poly(ADP-ribose) polymerase 1 in the response to oxidative stress, Mol. Cell. Biol., 25, 7625–7636, doi: https://doi.org/10.1128/MCB.25.17.7625-7636.2005.
Evdokimov, A. N., Petruseva, I. O., Pestryakov, P. E., and Lavrik, O. I. (2011) Photoactivated DNA analogs of substrates of the nucleotide excision repair system and their interaction with proteins of NER-competent extract of HeLa cells. Synthesis and application of long model DNA, Biochemistry (Moscow), 76, 157–166.
Yu, Y., and Waters, R. (2005) Histone acetylation, chromatin remodeling and nucleotide excision repair: hint from the study on MFA2 in Saccharomyces cerevisiae, Cell Cycle, 4, 1043–1045, doi: https://doi.org/10.4161/cc.4.8.1928.
Waters, R., van Eijk, P., and Reed, S. (2015) Histone modification and chromatin remodeling during NER, DNA Repair (Amst.), 36, 105–113, doi: https://doi.org/10.1016/j.dnarep.2015.09.013.
Yu, S., Evans, K., van Eijk, P., Bennett, M., Webster, R. M., Leadbitter, M., Teng, Y., Waters, R., Jackson, S. P., and Reed, S. H. (2016) Global genome nucleotide excision repair is organized into domains that promote efficient DNA repair in chromatin, Genome Res., 26, 1376–1387, doi: https://doi.org/10.1101/gr.209106.116.
Datta, A., Bagchi, S., Nag, A., Shiyanov, P., Adami, G. R., Yoon, T., and Raychaudhuri, P. (2001) The p48 subunit of the damaged-DNA binding protein DDB associates with the CBP/p300 family of histone acetyltransferase, Mutat. Res., 486, 89–97.
Rapic-Otrin, V., McLenigan, M. P., Bisi, D. C., Gonzalez, M., and Levine, A. S. (2002) Sequential binding of UV DNA damage binding factor and degradation of the p48 subunit as early events after UV irradiation, Nucleic Acids Res., 30, 2588–2598.
Martinez, E., Palhan, V. B., Tjernberg, A., Lymar, E. S., Gamper, A. M., Kundu, T. K., Chait, B. T., and Roeder, R. G. (2001) Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo, Mol. Cell. Biol., 21, 6782–6795, doi: https://doi.org/10.1128/MCB.21.20.6782-6795.2001.
Matsunuma, R., Niida, H., Ohhata, T., Kitagawa, K., Sakai, S., Uchida, C., Shiotani, B., Matsumoto, M., Nakayama, K. I., Ogura, H., Shiiya, N., and Kitagawa, M. (2015) UV damage-induced phosphorylation of HBO1 triggers CRL4DDB2-mediated degradation to regulate cell proliferation, Mol. Cell. Biol., 36, 394–406, doi: https://doi.org/10.1128/MCB.00809-15.
Zhao, R., Han, C., Eisenhauer, E., Kroger, J., Zhao, W., Yu, J., Selvendiran, K., Liu, X., Wani, A. A., and Wang, Q. E. (2014) DNA damage-binding complex recruits HDAC1 to repress Bcl-2 transcription in human ovarian cancer cells, Mol. Cancer Res., 12, 370–380, doi: https://doi.org/10.1158/1541-7786.MCR-13-0281.
Zhu, Q., Battu, A., Ray, A., Wani, G., Qian, J., He, J., Wang, Q. E., and Wani, A. A. (2015) Damaged DNA-binding protein down-regulates epigenetic mark H3K56Ac through histone deacetylase 1 and 2, Mutat. Res., 776, 16–23, doi: https://doi.org/10.1016/j.mrfmmm.2015.01.005.
Kakumu, E., Nakanishi, S., Shiratori, H. M., Kato, A., Kobayashi, W., Machida, S., Yasuda, T., Adachi, N., Saito, N., Ikura, T., Kurumizaka, H., Kimura, H., Yokoi, M., Sakai, W., and Sugasawa, K. (2017) Xeroderma pigmento-sum group C protein interacts with histones: regulation by acetylated states of histone H3, Genes Cells, 22, 310–327, doi: https://doi.org/10.1111/gtc.12479.
Tillhon, M., Cazzalini, O., Nardo, T., Necchi, D., Sommatis, S., Stivala, L. A., Scovassi, A. I., and Prosperi, E. (2012) p300/CBP acetyl transferases interact with and acetylate the nucleotide excision repair factor XPG, DNA Repair (Amst.), 11, 844–852, doi: https://doi.org/10.1016/j.dnarep.2012.08.001.
Fan, W., and Luo, J. (2010) SIRT1 regulates UV-induced DNA repair through deacetylating XPA, Mol. Cell, 39, 247–258, doi: https://doi.org/10.1016/j.molcel.2010.07.006.
Zhao, M., Geng, R., Guo, X., Yuan, R., Zhou, X., Zhong, Y., Huo, Y., Zhou, M., Shen, Q., Li, Y., Zhu, W., and Wang, J. (2017) PCAF/GCN5-mediated acetylation of RPA1 promotes nucleotide excision repair, Cell Rep., 20, 1997–2009, doi: https://doi.org/10.1016/j.celrep.2017.08.015.
He, H., Wang, J., and Liu, T. (2017) UV-Induced RPA1 acetylation promotes nucleotide excision repair, Cell Rep., 20, 2010–2025, doi: https://doi.org/10.1016/j.celrep.2017.08.016.
Matsuoka, S., Ballif, B. A., Smogorzewska, A., McDonald, E. R., 3rd, Hurov, K. E., Luo, J., Bakalarski, C. E., Zhao, Z., Solimini, N., Lerenthal, Y., Shiloh, Y., Gygi, S. P., and Elledge, S. J. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage, Science, 316, 1160–1166, doi: https://doi.org/10.1126/science.1140321.
Zannini, L., Delia, D., and Buscemi, G. (2014) CHK2 kinase in the DNA damage response and beyond, J. Mol. Cell. Biol., 6, 442–457, doi: https://doi.org/10.1093/jmcb/mju045.
Attwood, P. V., Besant, P. G., and Piggott, M. J. (2011) Focus on phosphoaspartate and phosphoglutamate, Amino Acids, 40, 1035–1051, doi: https://doi.org/10.1007/s00726-010-0738-5.
Hornbeck, P. V., Kornhauser, J. M., Tkachev, S., Zhang, B., Skrzypek, E., Murray, B., Latham, V., and Sullivan, M. (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse, Nucleic Acids Res., 40, D261–D270, doi: https://doi.org/10.1093/nar/gkr1122.
Shah, P., Zhao, B., Qiang, L., and He, Y. Y. (2018) Phosphorylation of xeroderma pigmentosum group C regulates ultraviolet-induced DNA damage repair, Nucleic Acids Res., 46, 5050–5060, doi: https://doi.org/10.1093/nar/gky239 https://doi.org/10.1093/nar/gky239.
Wu, X., Shell, S. M., Yang, Z., and Zou, Y. (2006) Phosphorylation of nucleotide excision repair factor xeroderma pigmentosum group A by ataxia telangiectasia mutated and Rad3-related-dependent checkpoint pathway promotes cell survival in response to UV irradiation, Cancer Res., 66, 2997–3005, doi: https://doi.org/10.1158/0008-5472.CAN-05-3403.
Lee, T. H., Park, J. M., Leem, S. H., and Kang, T. H. (2014) Coordinated regulation of XPA stability by ATR and HERC2 during nucleotide excision repair, Oncogene, 33, 19–25, doi: https://doi.org/10.1038/onc.2012.539.
Coin, F., Auriol, J., Tapias, A., Clivio, P., Vermeulen, W., and Egly, J. M. (2004) Phosphorylation of XPB helicase regulates TFIIH nucleotide excision repair activity, EMBO J., 23, 4835–4846, doi: https://doi.org/10.1038/sj.emboj.7600480.
Oakley, G. G., Loberg, L. I., Yao, J., Risinger, M. A., Yunker, R. L., Zernik-Kobak, M., Khanna, K. K., Lavin, M. F., Carty, M. P., and Dixon, K. (2001) UV-induced hyperphosphorylation of replication protein A depends on DNA replication and expression of ATM protein, Mol. Biol. Cell, 12, 1199–1213, doi: https://doi.org/10.1091/mbc.12.5.1199.
Liu, V. F., and Weaver, D. T. (1993) The ionizing irradiation-induced replication protein A phosphorylation response differs between ataxia telangiectasia and normal human cells, Mol. Cell. Biol., 13, 7222–7231.
Rodrigo, G., Roumagnac, S., Wold, M. S., Salles, B., and Calsou, P. (2000) DNA replication but not nucleotide excision repair is required for UVC-induced replication protein A phosphorylation in mammalian cells, Mol. Cell. Biol., 20, 2696–2705.
Niida, H., Matsunuma, R., Horiguchi, R., Uchida, C., Nakazawa, Y., Motegi, A., Nishimoto, K., Sakai, S., Ohhata, T., Kitagawa, K., Moriwaki, S., Nishitani, H., Ui, A., Ogi, T., and Kitagawa, M. (2017) Phosphorylated HBO1 at UV irradiated sites is essential for nucleotide excision repair, Nat. Commun., 8, 16102, doi: https://doi.org/10.1038/ncomms16102.
Krishnakumar, R., and Kraus, W. L. (2010) The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets, Mol. Cell, 39, 8–24, doi: https://doi.org/10.1016/j.molcel.2010.06.017.
Acknowledgements. The authors are grateful to Yu. S. Krasikova for help in the manuscript preparation.
Funding
Funding. The work was supported by the Russian Foundation for Basic Research (projects nos. 18-04-00596 and 19-04-00481) and the Program of Basic Scientific Research of the State Academies of Sciences for 2013–2020 (nos. AAAA-A17-117020210022-4 for O.I.L.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical norm compliance. This review does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Conflict of interest. The authors declare no conflict of interest.
Russian Text © The Author(s), 2019, published in Biokhimiya, 2019, Vol. 84, No. 9, pp. 1244–1258.
Rights and permissions
About this article
Cite this article
Rechkunova, N.I., Maltseva, E.A. & Lavrik, O.I. Post-translational Modifications of Nucleotide Excision Repair Proteins and Their Role in the DNA Repair. Biochemistry Moscow 84, 1008–1020 (2019). https://doi.org/10.1134/S0006297919090037
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297919090037