Abstract
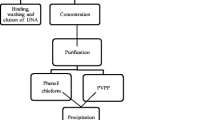
A new method for extracting DNA from plants is proposed, using the example of wild grapes Vitis amurensis Rupr. for further preparation of libraries for metagenomic analysis. The method is based on the isolation of DNA by an inexpensive CTAB method with an additional stage of DNA purification using silica spin columns (CTAB-spin method). A comparative analysis of the results of metagenomic analysis of endophytes on DNA isolated using the proposed CTAB-spin method and using the commercial kit ZymoBIOMICS DNA Miniprep (Zymo Research) was performed. It was found that when using the CTAB-spin method, the number of sequences of the 16S rRNA site and the diversity of bacterial genera were 2.8 and 1.2 times greater, respectively, than when using the ZymoBIOMICS kit. At the same time, the number of sequences of the internal transcribed spacer 1 (ITS1) and the biodiversity of endophytic fungi did not differ significantly during DNA extraction by two methods. Thus, the proposed method of DNA isolation for metagenomic analysis is an available and effective alternative to commercial kits for the isolation of plant DNA for new-generation sequencing methods.


Similar content being viewed by others
REFERENCES
Behjati, S. and Tarpey, P.S., ADS—Educ. Pract., 2013, vol. 98, pp. 236–238.
Slatko, B.E., Gardner, A.F., and Ausubel, F.M., Curr. Protoc. Mol. Biol., 2018, vol. 122, p. e59. https://doi.org/10.1002/cpmb.59
Kulski, J.K., Next-Generation Sequencing—An Overview of the History, Tools, and “Omic” Applications, Kulski, J.K., Ed., IntechOpen, 2016, p. 60. https://doi.org/10.5772/61964
Lam, H.Y.K., Clark, M.J., Chen, R., Chen, R., Natsoulis, G., O’Huallachain, M., et al., Nat. Biotechnol., 2012, vol. 30, pp. 78–82.
Wang, Z., Gerstein, M., and Snyder, M., Nat. Rev. Genet., 2009, vol. 10, pp. 57–63.
Rabbani, B., Tekin, M., and Mahdieh, N., J. Hum. Genet., 2014, vol. 59, pp. 5–15.
Leo, V.C., Morgan, N.V., Bem, D., Jones, M.L., Lowe, G.C., Lordkipanidze, M., et al., J. Thromb. Haemostas., 2015, vol. 13, pp. 643–650.
Kulski, J.K., Suzuki, S., Ozaki, Y., Mitsunaga, S., Inoko, H., and Shiina, T., Phase HLA Genotyping by NGS—A Comparison Between two Massively Parallel Sequencing Bench-top Systems, the Roche GS Junior and Ion Torrent PGM, Xi, Y., Ed., IntechOpen, 2014, pp. 141–181.
Pelizzola, M. and Ecker, J.R., FEBS Lett., 2011, vol. 585, pp. 1994–2000.
Simner, P.J., Miller, S., and Carroll, K.C., Clin. Infect. Dis., 2018, vol. 66, pp. 778–788.
Boers, S.A., Jansen, R., and Hays, J.P., Eur. J. Clin. Microbiol. Infect. Dis., 2019, vol. 38, pp. 1059–1070.
Chiu, C.Y. and Miller, S.A., Nat. Rev. Genet., 2019, vol. 20, pp. 341–355.
Iquebal, M.A., Jagannadham, J., Jaiswal, S., Prabha, R., Rai, A., and Kumar, D., Front. Microbiol., 2022, vol. 13, p. 708335. https://doi.org/10.3389/fmicb.2022.708335
Fan, Y., Gao, L., Chang, P., and Li, Z., Ann. Microbiol., 2020, vol. 70, p. 30. https://doi.org/10.1186/s13213-020-01574-9
Cureau, N., Threlfall, R., Marasini, D., Lavefve, L., and Carbonero, F., Microbiol. Ecol., 2021, vol. 82, pp. 845–858.
Marasco, R., Rolli, E., Fusi, M., Michoud, G., and Daffonchio, D., Microbiome, 2018, vol. 6, p. 3. https://doi.org/10.1186/s40168-017-0391-2
Deyett, E. and Rolshausen, P.E., Front. Plant Sci., 2019, vol. 10, p. 1246. https://doi.org/10.3389/fpls.2019.01246
Kiselev, K.V., Tyunin, A.P., and Karetin, Y.A., Plant Cell Rep., 2015, vol. 34, pp. 311–320.
Ogneva, Z.V., Dubrovina, A.S., and Kiselev, K.V., Biol. Plant., 2016, vol. 60, pp. 628–634.
Aleynova, O.A., Nityagovsky, N.N., Dubrovina, A.S., and Kiselev, K.V., Plants, 2022, vol. 11, p. 1128. https://doi.org/10.3390/plants10071276
Deyett, E. and Rolshausen, P.E., FEMS Microbiol. Ecol., 2020, vol. 96, p. fiaa053. https://doi.org/10.1093/femsec/fiaa053
Bolyen, E., Rideout, J.R., Dillon, M.R., Bokulich, N.A., and Abnet, C.C., Al-Ghalith, M., et al., Nat. Biotechnol., 2019, vol. 37, pp. 852–857.
Callahan, B.J., McMurdie, P.J., Rosen, M.J., Han, A.W., Johnson, A.J.A., and Holmes, S.P., Nat. Methods, 2016, vol. 13, pp. 581–583.
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., et al., J. Machine Learn. Res., 2011, vol. 12, pp. 2825–2830.
Bokulich, N.A., Kaehler, B.D., Rideout, J.R., Dillon, M., Bolyen, E., et al., Microbiome, 2018, vol. 6, p. 90. https://doi.org/10.1186/s40168-018-0470-z
Nilsson, R.H., Larsson, K.-H., Taylor, A.F.S., Bengtsson-Palme, J., Jeppesen, T.S., Schigel, D., et al., Nucleic Acids Res., 2019, vol. 47, pp. D259–D264.
McMurdie, P.J. and Holmes, S., PLoS One, 2013, vol. 8, p. e61217. https://doi.org/10.1371/journal.pone.0061217
Wickham, H., Averick, M., Bryan, J., Chang, W., McGowan, L.D., Francois, R., et al., J. Open Source Software, 2019, vol. 4, p. 1686. https://doi.org/10.21105/joss.01686
Oksanen, J., Blanchet, F.G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al., Vegan: Community Ecology Package, R Package Version 2.5-7. 2020. https://cran.r-project.org/web/packages/vegan/vegan.pdf. Accessed January 9, 2023.
Gu, Z., Eils, R., and Schlesner, M., Bioinformatics, 2016, vol. 32, pp. 2847–2849.
Funding
The research was supported by the Russian Science Foundation, project no. 22-74-10001, https://rscf.ru/project/22-74-10001.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by E. Makeeva
Rights and permissions
About this article
Cite this article
Kiselev, K.V., Nityagovsky, N.N. & Aleynova, O.A. A Method of DNA Extraction from Plants for Metagenomic Analysis Based on the Example of Grape Vitis amurensis Rupr.. Appl Biochem Microbiol 59, 361–367 (2023). https://doi.org/10.1134/S0003683823030110
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0003683823030110