Abstract
The muscular ventricular septum separates the flow of oxygenated and de-oxygenated blood in air-breathing vertebrates. Defects within it, termed muscular ventricular septal defects (VSDs), are common, yet less is known about how they arise than rarer heart defects. Mutations of the cardiac transcription factor NKX2-5 cause cardiac malformations, including muscular VSDs. We describe here a genetic interaction between Nkx2-5 and Sarcospan (Sspn) that affects the risk of muscular VSD in mice. Sspn encodes a protein in the dystrophin-glycoprotein complex. Sspn knockout (SspnKO) mice do not have heart defects, but Nkx2-5+/−/SspnKO mutants have a higher incidence of muscular VSD than Nkx2-5+/− mice. Myofibers in the ventricular septum follow a stereotypical pattern that is disrupted around a muscular VSD. Subendocardial myofibers normally run in parallel along the left ventricular outflow tract, but in the Nkx2-5+/−/SspnKO mutant they commonly deviate into the septum even in the absence of a muscular VSD. Thus, Nkx2-5 and Sspn act in a pathway that affects the alignment of myofibers during the development of the ventricular septum. The malalignment may be a consequence of a defect in the coalescence of trabeculae into the developing ventricular septum, which has been hypothesized to be the mechanistic basis of muscular VSDs.
Similar content being viewed by others
Introduction
Muscular VSDs are anatomically simple, but their pathogenesis is ironically less well understood than more complex heart defects such as tetralogy of Fallot. Two general hypotheses, extrapolated from observations of normal cardiac development in the chick embryo, were proposed decades ago to explain how a muscular VSD arises. Early observations of cell death suggested that excessive apoptosis could perforate the septum1,2. Increased myocyte apoptosis is not typically observed in association with abnormal ventricular septal development, however, even if the genetic perturbation is severe3. Alternatively, the ventricular septum in chick embryos forms by the coalescence of trabeculae4,5,6. Muscular VSDs were hence proposed to represent the persistence of gaps between trabeculae during their coalescence5.
The development of the muscular ventricular septum in mouse models has not been scrutinized as closely as other cardiac structures, but morphologic observations support the role of trabecular coalescence7,8. Certain mutant phenotypes suggest that the process of coalescence shares genetic pathways with trabecular development. For example, conditional knockout mutations that disrupt Notch signaling cause hypertrabeculation and ventricular noncompaction phenotypes similar to those observed in humans. Investigators have thus focused their attention upon the ventricular myocardium of the free wall, but they have made note of muscular and membranous VSDs9,10,11. Curiously, the ventricular septum in the mutants is not necessarily hypertrabeculated or thin like the compact myocardium of the free wall. Some regions of the ventricular septum can have normal thickness and shape, while gaps elsewhere suggest a local failure of trabeculae to coalesce (cf. Fig. 6 in ref. 8, Fig. 1a in ref. 9, Fig. 4a in ref. 10, and Figs 2 and 7 in ref. 12, which show examples from cardiomyocyte-specific knockouts of Mib18,9, Fkbp1a10, and Scrib and Rac112).
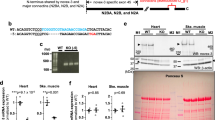
(a) SSPN is a transmembrane protein in the dystrophin-glycoprotein complex. The highly conserved glutamine (Q) at position 213 of mouse SSPN resides in the C-terminal cytoplasmic domain. (b) Q213 is nearly invariant across Sarcopterygii species. C57BL/6 carries a missense mutation (Q213L), whereas FVB/N carries the conserved glutamine. The multispecies alignment in the C-terminal portion of the protein is shown for representative Sarcopterygii and Actinopterygii species. Mouse position 213 is indicated. Amino acid conservation is scored separately for the Sarcopterygii and Actinopterygii. The maximum score, 11, corresponds to perfect conservation of an amino acid across all species. The full-length sequences of additional species are aligned in Supplementary Fig. 1. (c) The expression of Sspn mRNA in the heart is the same across the inbred C57BL/6N (N = 3 per genotype), FVB/N (N = 3) and C57BL/6N X FVB/N F1 hybrid backgrounds (N = 6 per genotype) and wild-type and Nkx2-5+/− genotypes. No transcript is detected in SspnKO hearts (N = 2). Mean ± S.D. are shown.
The left panel of each image pair shows the ventricular septum of representative wild-type (N = 5), SspnKO (N = 4) and Nkx2-5+/− (N = 3) single mutants, and double mutant Nkx2-5+/−/SspnKO (N = 4) hearts. The yellow vectors indicate the orientation of fibers that run parallel to the plane of section. The right panel shows the orientation of myofibers within the boxed region. Subendocardial myofibers on the left side of the ventricular septum generally run in parallel from the apex to base in every wild-type and single mutant heart examined. In each of the double mutant Nkx2-5+/−/SspnKO hearts, the left-sided subendocardial fibers take abnormal turns toward the right (arrowheads), sometimes forming whorls. The arrows point to two whorls. None of the hearts examined has a muscular VSD. Scale bar, 200 μm.
(a) Nkx2-5+/− mice have an increased incidence of ASD and membranous and muscular VSD compared to the Nkx2-5WT, regardless of the Sspn genotype. SspnWT and Sspn+/− genotypes are combined because their separate incidences of any defect are the same regardless of Nkx2-5 genotype. The SspnKO genotype increases the incidence of muscular VSD in the Nkx2-5+/− background but not ASD or membranous VSD. The SspnKO mutation alone does cause heart defects. N = 217 Nkx2-5WT/SspnWT & /Sspn+/−, 106 Nkx2-5WT/SspnKO, 301 Nkx2-5+/−/SspnWT & /Sspn+/−, 161 Nkx2-5+/−/SspnKO. *P < 0.05. **P < 0.01.
If the conditional knockout mutants above represent extreme examples of how muscular VSDs arise, then subtler signs of abnormal trabecular coalescence may be associated with milder mutations of cardiac developmental genes that cause muscular VSD. Among the genes, mutations of NKX2-5, a cardiac transcription factor, cause pleiotropic congenital heart defects13,14. The anatomic phenotype is determined in part by modifier genes that influence the susceptibility of specific developmental pathways to Nkx2-5 mutation14,15. One modifier locus on chromosome 6 for muscular VSDs was previously mapped in a cross between the C57BL/6N and FVB/N inbred strains15. The paucity of knowledge regarding how a muscular VSD arises motivated us to evaluate sarcospan, a gene in the locus.
Sarcospan (SSPN) is a transmembrane protein in the dystrophin-glycoprotein complex. The complex links the extracellular matrix to the membrane cytoskeleton of myocytes. Sspn is specifically expressed in the heart and skeletal muscle16,17. Mutations of dystrophin-glycoprotein complex genes cause muscular dystrophy. Mutations are not usually associated with congenital heart disease, although an alpha-dystrobrevin mutation in one family was associated with VSD, hypoplastic left heart, and left ventricular noncompaction18. C57BL/6N, which bears the risk allele at the chromosome 6 locus, carries a missense mutation of a highly conserved glutamine at position 213 of SSPN. The amino acid is in the cytoplasmic C-terminal domain (Fig. 1a). Sspn knockout (SspnKO) mice have normal skeletal muscle function. The null mutant is obtained at the expected Mendelian frequency19, so Sspn deficiency alone would not be expected to cause serious congenital heart defects.
Results
Identification of Sarcospan as a candidate modifier gene for muscular VSD in Nkx2-5+/− mice
We sequenced the Sspn alleles carried by the parental strains in our mapping cross. We confirmed that FVB/N carries the conserved glutamine (Q) and C57BL/6N carries a leucine (L) at position 213 (Fig. 1b and Supplementary Fig. 1). The function of the conserved glutamine at position 213, which aligns to position 240 in humans, is unknown, but a survey of human and other vertebrate species suggests that the amino acid was strongly selected at the divergence of lobe- and ray-finned fishes. All 65,777 humans in the 1000 Genomes Project20, NHLBI GO Exome Sequencing Project21, and the Exome Aggregation Consortium22 and nearly all Sarcopterygii species, e.g., coelacanth, lungfish and tetrapods, carry a glutamine at the position mutated in C57BL/6 (Fig. 1b and Supplementary Fig. 1). The equivalent Q240L mutation in human SSPN has a scaled CADD score of 12.4, placing it in the top 10% of potential variants ranked by predicted deleteriousness23. Actinopterygii, i.e., ray-finned fishes, all carry a tryptophan (W) at the same position (Fig. 1b and Supplementary Fig. 1). Q240W has a scaled CADD score of 25, placing it in the top 1% of predicted deleterious variants23.
Some chromatin immunoprecipitation data suggest that NKX2-5 binds to potential Sspn regulatory elements24,25, but we found no evidence for a regulatory polymorphism that affects expression. Sspn mRNA levels are the same in the hearts of C57BL/6N, FVB/N and C57BL/6N X FVB/N F1 hybrid mice. Wild-type and Nkx2-5+/− mice also express Sspn at the same level (Fig. 1c).
The Sspn null mutation increases the incidence of muscular VSD in Nkx2-5+/− mice
We next investigated whether Sspn genetically interacts with Nkx2-5 in the development of the muscular ventricular septum. Double heterozygotes, i.e., Nkx2-5+/−/Sspn+/−, were crossed to each other or Sspn+/− single mutants. The Nkx2-5 null mutation is embryonic lethal26,27, but all other combinations of Nkx2-5 and Sspn genotypes were recovered at the expected Mendelian frequency in newborn mouse pups (Supplementary Fig. 2). The co-occurrence of Nkx2-5+/− and a heterozygous or homozygous Sspn knockout mutation does not affect embryonic survival.
The null mutation of Sspn (SspnKO) does not cause congenital heart defects in Nkx2-5 wild-type (Nkx2-5WT) mice (Fig. 2). The incidences of each type of septal defect among the Sspn wild-type (SspnWT) and Sspn+/− mice in either the Nkx2-5+/− or Nkx2-5WT groups were the same, so their results were combined for statistical analyses.
As expected, Nkx2-5+/− mice have an increased incidence of atrial septal defects (ASDs), membranous VSDs and muscular VSDs compared to the Nkx2-5WT. The SspnKO genotype does not affect the incidences of ASD or membranous VSD in the Nkx2-5+/− mice, but Nkx2-5+/−/SspnKO double mutants have almost twice the incidence of muscular VSD compared to Nkx2-5+/− single mutants (Fig. 2). The results place Sspn in a genetic pathway that leads from Nkx2-5 mutation to a defect in the muscular ventricular septum.
Incidentally, a low incidence of muscular VSD was observed in Nkx2-5WT hearts. In a previous study of Nkx2-5WT mice in the C57BL/6N strain, which is also the predominant background of the mice in this study, we did not detect the <5% incidence probably because of a smaller sample size14. The prior study did note, however, a correlation between muscular VSD incidence in Nkx2-5+/− mice and the fraction of C57BL/6N genome in the genetic background of inbred strain crosses with FVB/N and A/J. C57BL/6N may carry genetic polymorphisms that increase the propensity toward muscular VSD in Nkx2-5WT hearts as well.
The Sspn null mutation disrupts the organization of myocytes in the ventricular septum of Nkx2-5+/− mice
The inspection of thousands of hearts in several projects led us to notice that the myofibers around a muscular VSD veer from their normal trajectories. For example, in a heart with an intact ventricular septum the subendocardial fibers on the left side of the septum tend to run in parallel from the apex to the base (Fig. 3a). In a heart with a muscular VSD, the left-sided, subendocardial fibers veer toward the right in sections that are near the defect (Fig. 3b). Fibers that surround the defect appear even more malaligned (Fig. 3c). The malalignment could either be a simple consequence of myofiber diversion around a defect or a sign of flawed trabecular coalescence, which could be the basis of a muscular VSD. To distinguish the two possibilities, we cut 1 μm, thin sections of plastic-embedded hearts. The method preserves the myofiber architecture better than paraffin embedding, which is more amenable to sectioning large number of hearts quickly.
(a) In an Nkx2-5+/− heart with an intact ventricular septum, the subendocardial fibers on the left side of the septum run parallel to the outflow tract, as indicated by the arrow. (b,b’) In an Nkx2-5+/− heart with a muscular VSD, the left-sided subendocardial fibers deviate toward the right and split in two directions in the region just ventral to the VSD. One branch continues to the right side. At the level of the VSD, the fibers are generally malaligned. The upper and lower panels are shown at 5x and 20x magnification, respectively. RV, LV, right and left ventricle.
The orientation of myofibers in the normal heart follows a stereotypical pattern28. In the left ventricular free wall, subepicardial fibers run with a major component parallel to the long axis from apex to base, while fibers in the middle layer run circumferentially around the ventricle. The left ventricular free wall in the four genotypic groups – wild type, Nkx2-5+/− and SspnKO single mutants, and Nkx2-5+/−/SspnKO double mutant – showed this pattern (Supplementary Fig. 3).
In the ventricular septum, the subendocardial fibers run in parallel along the left ventricular outflow tract. Subendocardial fibers on the right ventricular side do not have as consistent a pattern, perhaps owing to the shape of the right ventricle relative to the plane of section. Fibers in the middle layer of the septum run circumferentially. This pattern was observed in the intact ventricular septum of every wildtype and Nkx2-5+/− and SspnKO single mutant heart (Fig. 4). In contrast, each of the Nkx2-5+/−/SspnKO double mutants examined had a subset of subendocardial fibers on the left side of the ventricular septum that deviated toward the right, sometimes forming whorls, even though there was no VSD (Fig. 4 and Supplementary Fig. 4). The incidence of abnormal myofiber organization in the Nkx2-5+/−/SspnKO double mutant is significantly greater than in the other three genotypic groups (P < 0.001, two-tailed Fisher exact test). An abnormal course of myofibers in the ventricular septum is not simply the consequence of a muscular defect. Collectively, the results indicate that Nkx2-5 and Sspn operate together in a pathway that affects the alignment of myofibers in the ventricular septum.
Discussion
Dozens of genetic mutations are known to cause muscular VSDs, but how the heart defect forms is poorly understood29,30. A novel genetic interaction between Nkx2-5 and Sspn offers insight into the mechanism. NKX2-5 regulates diverse cardiac developmental pathways13. Polymorphisms of modifier genes in these pathways influence their susceptibility to Nkx2-5 haploinsufficiency and the expression of the mutant phenotype14. Sspn is a gene that resides in a chromosome 6 locus that modifies the risk of muscular VSD in Nkx2-5+/− mice15. The present data indicate that Sspn is an Nkx2-5 modifier gene, although we cannot state definitively whether it is the gene that underlies the chromosome 6 locus. Complete loss of Sspn function does not cause heart defects but does increase the incidence of muscular VSD in combination with an Nkx2-5 mutation. In addition, myofibers in the ventricular septum of Nkx2-5+/−/SspnKO double mutants commonly deviate from their normal trajectories. Myofibers similarly deviate around muscular VSDs, as noticed by us and another group31. We propose that the malaligned septal myofibers are a sign of a defect in trabecular coalescence that causes muscular VSDs.
The development of the muscular ventricular septum begins with the coalescence of trabeculations in the interventricular groove of the looping heart tube6,7. Lineage analyses in the mouse demonstrate that left and right ventricular myocytes form their respective sides of the septum. A sharp, left-right boundary delineates their contributions to the septum32. Myofibers that run from left to the right side of the ventricular septum appear to violate this boundary. Certain defects of trabecular development could disrupt their coalescence into the ventricular septum, thus causing the malalignment of myofibers or a muscular VSD. For example, cardiomyocyte-specific deletion of Scrib, a planar cell polarity gene, causes the stunting of trabeculae. Scrib localizes the GTPase signaling protein Rac1 to the cell membrane. Their interaction plays a critical role in the organization of trabecular myocytes, the apposition of myocytes in the ventricular septum, and the regulation of junctional complex formation at the cell membrane12. A quantitative defect in these processes could cause a trabecula to become malaligned if it has no neighboring trabeculae to support its normal coalescence. A muscular VSD could result when malaligned trabeculae do not come into apposition.
Interestingly, there may be subtle, quantitative differences in trabecular developmental processes even between wild-type animals. Fractal analyses indicate that the morphologic pattern of the trabecular network is more complex in the inbred C57BL/6 strain compared to the outbred NIMR:Parkes8. This complex, but quantifiable trabecular phenotype may be related to the low incidence of muscular VSDs in Nkx2-5WT mice from a predominantly C57BL/6N background. Fractal analyses of trabeculae, as described by Captur et al.8,33, in mutants prone to muscular VSD could help to illuminate patterns of trabecular coalescence during cardiac development and the functional significance of myofiber malalignment.
The molecular and cellular bases of the genetic interaction between Nkx2-5 and Sspn remain to be defined. The hypertrabeculation phenotypes associated with human and mouse mutations indicate that NKX2-5 plays a role in trabecular development34,35. The present results indicate that loss of Sspn exacerbates a subtle defect associated with Nkx2-5 haploinsufficiency. In light of what is known about dystrophin-associated complex genes in skeletal muscle and other organisms, one can consider potential physical or biological functions of Sspn in ventricular septal development. The dystrophin-glycoprotein complex is best known for its role in maintaining the integrity of the muscle cell membrane and transducing force36. Mutations that cause muscular dystrophy are not associated with congenital heart defects, so a purely mechanical function of Sspn in ventricular septal development seems less likely. The dystrophin-glycoprotein complex also plays a role in signaling. SSPN promotes Akt signaling during muscle regeneration, and the dystrophin-glycoprotein complex modulates integrin signaling37,38,39. Elegant studies in Drosophila clearly demonstrate the functions of the dystrophin-associated complex in regulating planar cell polarity40,41, BMP and Notch signaling42,43. All these pathways are all involved in the formation of the ventricular myocardium11, and NKX2-5 has been shown to regulate BMP and Notch signaling44,45.
The function of amino acid Q213 and the effect of the Q213L mutation on Sspn function are unknown. The cytoplasmic C-terminal domain of SSPN, which includes Q213, contributes to protein-protein interactions that stabilize the dystrophin-glycoprotein complex46. The amino acid could function in this capacity or in a different protein-protein interaction. Interestingly, the conserved glutamine arose in the Sarcopterygii lineage in association with the onset of the development of a ventricular septum. The African lungfish, an early Sarcopterygii species, has a rudimentary ventricular septum47, whereas ray-finned fishes do not. Sspn is not essential for the formation of a ventricular septum, but the evolution of the ventricular septum undoubtedly involved the selection of polymorphisms present in the lobe-finned fishes.
Much remains to be learned regarding how mutations of genes like Nkx2-5 and their interactions with modifier genes cause muscular VSDs. Although muscular VSDs are usually benign, the investigation of their pathogenesis should illuminate mechanisms pertinent to congenital heart disease, cardiomyopathy and cardiac tissue engineering.
Methods
Mouse strains
Sspn knockout mice were purchased from the Jackson Laboratory. They were predominantly in a C57BL/6 background. The Sspn knockout was made in 129S1/Sv x 129X1/SvJ F1 ES cells and backcrossed to C57BL/6 for five generations prior to arrival at Jackson. The Sspn wild-type and knockout alleles were genotyped as described19. Nkx2-5+/− mice were maintained in the C57BL/6N background and genotyped as previously described14,27. The C57BL/6N and FVB/N inbred strains were purchased from Charles River. The experiments were approved by the animal studies committee at Washington University School of Medicine and performed in accordance with their guidelines.
Sarcospan sequence analyses
SNPs among inbred mouse strains in Sspn were identified through a search of Mouse Genome Informatics (http://www.informatics.jax.org/strains_SNPs.shtml). The sequences of Sspn for the C57BL/6N and FVB/N strains in our colony were confirmed by cloning cDNA and sequencing on an Applied Biosystems 3730 DNA analyzer.
SNPs of human SSPN were queried on August 17, 2016 in the following databases: 1000 Genomes20 (N = 2504 individuals), Exome Aggregation Consortium22 (N = 60706), and NHLBI GO Exome Sequencing Project21 (N = 6503). The Exome Aggregation Consortium database includes the data of 3936 individuals from the NHLBI GO Exome Sequencing Project.
The reference sequences of SSPN from multiple species were downloaded from Ensembl and NCBI on April 15, 2015. Sequences that lacked the C-terminus or the expected stop codon or had large gaps (≥10 amino acids) were excluded from multi-species alignment analysis. The amino acid sequences of 48 species were aligned in Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/)48. The physiochemical conservation scores at each amino acid position was calculated and visualized in Jalview (http://www.jalview.org/)49. The transmembrane protein structure of mouse SSPN was predicted with the TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) and drawn with TMRPres2D (http://bioinformatics.biol.uoa.gr/TMRPres2D/)50,51.
The deleteriousness of an amino acid substitution was estimated on the human SSPN sequence by the Combined Annotation-Dependent Depletion (CADD) method23. Scaled CADD scores for codon substitutions at human Q240 of SSPN were obtained from the CADD website http://cadd.gs.washington.edu/home.
Quantification of Sspn mRNA expression
Total RNA was purified from C57BL/6N and FVB/N hearts using the RNeasy kit (Qiagen). cDNA was synthesized with a SuperScript Vilo cDNA synthesis kit (Invitrogen). Quantitative RT-PCR was performed using SYBR GreenER qPCR SuperMix (Invitrogen) on an Mx3005 P qPCR system (Stratagene). Sspn expression levels were normalized to Gapdh.
Phenotyping of congenital heart defects
Nkx2-5+/− mice were crossed to Sspn+/− or SspnKO mice to generate double heterozygote F1 offspring, i.e., Nkx2-5+/−/Sspn+/−, as well as the other genotypes. Nkx2-5+/−/Sspn+/− mice were then crossed to other double heterozygotes or Sspn+/− single mutants. The F2 offspring were collected within hours of birth. Their hearts were dissected, fixed in 10% formalin and processed for histology. The hearts were sectioned completely at 6 μm thickness and stained with hematoxylin and eosin. All the sections were examined for congenital heart defects without knowledge of genotype, as previously described14.
Assessment of cardiac myofiber orientation
Newborn mouse hearts were fixed in modified Karnovsky’s fixative (2% paraformaldehyde, 2.5% glutaraldehyde, 0.1 M cacodylate buffer, pH 7.4) and osmium tetroxide. The hearts were embedded in resin. Serial sections through the entire heart were cut at 1 μm thickness in the frontal plane and stained with toluidine blue. Sections were imaged with a Nanozoomer 2.0-HT digital slide scanner (Hamamatsu) at 20x magnification.
The orientation of myofibers was analyzed using ImageJ (http://fiji.sc) and an ImageJ plugin, OrientationJ (http://bigwww.epfl.ch/demo/orientation/)52. OrientationJ imposes a square grid on an image. The dominant orientation of pixels within each square is depicted as a vector. The analyses were performed by two individuals blinded to genotype.
Statistical analyses.
The incidences of a heart defect in two groups were compared in a 2 × 2 contingency table. Statistical significance was assessed by a chi-squared test.
The incidences of abnormal myofiber orientation were compared in a 2 × 4 contingency table. Statistical significance was assessed by a two-tailed, Fisher exact test.
Additional Information
How to cite this article: Panzer, A. A. et al. Nkx2-5 and Sarcospan genetically interact in the development of the muscular ventricular septum of the heart. Sci. Rep. 7, 46438; doi: 10.1038/srep46438 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
E. B. Clark . “Mechanisms in the Pathogenesis of Congenital Cardiac Malformations”. In Genetics of Cardiovascular Disease edited by M. E. Pierpont & J. H. Moller, pp. 3–11 (Martinus Nijhoff Publishing, Boston, 1987).
T. Pexieder . “Cell death in the morphogenesis and teratogenesis of the heart”. Adv. Anat. Embryol. Cell Biol. 51, 3–99 (1975).
J. K. Takeuchi et al. “Tbx5 specifies the left/right ventricles and ventricular septum position during cardiogenesis”. Development 130(24), 5953–5964 (2003).
J. Y. Harh & M. H. Paul . “Experimental cardiac morphogenesis. I. Development of the ventricular septum in the chick”. J. Embryol. Exp. Morphol. 33, 13–28 (1975).
G. Ben-Shachar et al. “Ventricular trabeculations in the chick embryo heart and their contribution to ventricular and muscular septal development”. Circ. Res. 57, 759–766 (1985).
A. Contreras-Ramos et al. “Normal development of the muscular region of the interventricular septum-I. The significance of the ventricular trabeculations”. Anat. Histol. Embryol. 37, 344–351 (2008).
R. H. Anderso et al. “The development of septation in the four-chambered heart”. Anat. Rec. (Hoboken) 297, 1414–1429 (2014).
G. Captur et al. “Morphogenesis of myocardial trabeculae in the mouse embry”. J. Anat. 229, 314–325 (2016).
G. Luxan et al. “Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy”. Nat. Med. 19, 193–201 (2013).
H. Chen et al. “Fkbp1a controls ventricular myocardium trabeculation and compaction by regulating endocardial Notch1 activity”. Development. 140, 1946–1957 (2013).
W. Zhang et al. “Molecular mechanism of ventricular trabeculation/compaction and the pathogenesis of the left ventricular noncompaction cardiomyopathy (LVNC)”. Am. J. Med Genet C. Semin. Med Genet. 163C, 144–156 (2013).
V. Boczonadi et al. “Scrib:Rac1 interactions are required for the morphogenesis of the ventricular myocardium”. Cardiovasc. Res. 104, 103–115 (2014).
D. W. Benson et al. “Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways”. J. Clin. Invest 104, 1567–1573 (1999).
J. B. Winston et al. “Heterogeneity of genetic modifiers ensures normal cardiac development”. Circulation 121, 1313–1321 (2010).
J. B. Winston et al. “Complex Trait Analysis of Ventricular Septal Defects Caused by Nkx2-5 Mutation”. Circ. Cardiovasc. Genet. 5, 293–300 (2012).
R. H. Crosbie et al. “Sarcospan, the 25-kDa transmembrane component of the dystrophin-glycoprotein complex”. J. Biol. Chem. 272, 31221–31224 (1997).
I. B. Chen et al. “Association of genes with physiological functions by comparative analysis of pooled expression microarray data”. Physiol Genomics 45, 69–78 (2013).
F. Ichida et al. “Novel gene mutations in patients with left ventricular noncompaction or Barth syndrome”. Circulation 103, 1256–1263 (2001).
C. S. Lebakken et al. “Sarcospan-deficient mice maintain normal muscle function”. Mol. Cell Biol. 20, 1669–1677 (2000).
A. Auton et al. “A global reference for human genetic variation”. Nature 526, 68–74 (2015).
Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/) [8-17-2015 accessed].
M. Lek et al. “Analysis of protein-coding genetic variation in 60,706 humans”. Nature 536, 285–291 (2016).
M. Kircher et al. “A general framework for estimating the relative pathogenicity of human genetic variants”. Nat. Genet. 46, 310–315 (2014).
R. Bouveret et al. “NKX2-5 mutations causative for congenital heart disease retain functionality and are directed to hundreds of targets”. Elife. 4, doi: 10.7554/eLife.06942. (2015).
A. He et al. “Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart”. Proc. Natl. Acad. Sci. USA. 108, 5632–5637 (2011).
I. Lyons et al. “Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5”. Genes Dev. 9(13), 1654–1666 (1995).
M. Tanaka et al. “The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development”. Development 126, 1269–1280 (1999).
D. D. Streeter, Jr. et al. “Fiber orientation in the canine left ventricle during diastole and systole”. Circ. Res. 24, 339–347 (1969).
K. Bellmann, A. Perrot & S. R. Sperling . “Human Genetics of Ventricular Septal Defect”. In Congenital Heart Diseases: The Broken Heart, edited by S. R. Sperling, R. G. Kelly & D. J. Driscoll, pp., 307–328 (Springer, Vienna, 2016).
L. Houyel . “Molecular Pathways and Animal Models of Ventricular Septal Defect’’. In Congenital Heart Diseases: The Broken Heart, edited by S. R. Sperling, R. G. Kelly & D. J. Driscoll, pp., 329–341 (Springer, Vienna, 2016)
A. L. McGinley et al. “Additional sex combs-like family genes are required for normal cardiovascular development”. Genesis. 52(7), 671–686 (2014).
D. Franco et al. “Left and right ventricular contributions to the formation of the interventricular septum in the mouse heart”. Dev. Biol 294, 366–375 (2006).
G. Captur et al. “Abnormal cardiac formation in hypertrophic cardiomyopathy: fractal analysis of trabeculae and preclinical gene expression”. Circ. Cardiovasc. Genet. 7, 241–248 (2014).
M. Pashmforoush et al. “Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block”. Cell 117(3), 373–386 (2004).
H. Ashraf et al. “A mouse model of human congenital heart disease: high incidence of diverse cardiac anomalies and ventricular noncompaction produced by heterozygous Nkx2-5 homeodomain missense mutation”. Circ. Cardiovasc. Genet. 7, 423–433 (2014).
F. Rahimov & L. M. Kunkel . “The cell biology of disease: cellular and molecular mechanisms underlying muscular dystrophy”. J. Cell Biol. 201, 499–510 (2013).
B. Constantin . “Dystrophin complex functions as a scaffold for signalling proteins”. Biochim. Biophys. Acta 1838, 635–642 (2014).
J. L. Marshall et al. “Sarcospan-dependent Akt activation is required for utrophin expression and muscle regeneration”. J. Cell Biol. 197, 1009–1027 (2012).
M. S. Parvatiyar et al. “Sarcospan Regulates Cardiac Isoproterenol Response and Prevents Duchenne Muscular Dystrophy-Associated Cardiomyopathy”. J. Am. Heart Assoc. 4, e002481 (2015).
V. Mirouse et al. “Dystroglycan and perlecan provide a basal cue required for epithelial polarity during energetic stress”. Dev. Cell. 16(1), 83–92 (2009).
W. M. Deng et al. “Dystroglycan is required for polarizing the epithelial cells and the oocyte in Drosophila”. Development. 130, 173–184 (2003).
M. M. Kucherenko et al. “Genetic modifier screens reveal new components that interact with the Drosophila dystroglycan-dystrophin complex”. PLoS. One. 3, e2418 (2008).
C. P. Christoforou et al. “The detached locus encodes Drosophila Dystrophin, which acts with other components of the Dystrophin Associated Protein Complex to influence intercellular signalling in developing wing veins”. Dev. Biol. 313, 519–532 (2008).
Y. Nakashima et al. “Nkx2-5 suppresses the proliferation of atrial myocytes and conduction system”. Circ. Res. 114, 1103–1113 (2014).
O. W. Prall et al. “An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation”. Cell 128, 947–959 (2007).
G. Miller et al. “Structural and functional analysis of the sarcoglycan-sarcospan subcomplex”. Exp. Cell Res. 313, 639–651 (2007).
J. M. Icardo et al. “Ventricle and outflow tract of the African lungfish Protopterus dolloi”. J. Morphol. 265, 43–51 (2005).
F. Sievers et al. “Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega”. Mol. Syst. Biol. 7, 539 (2011).
A. M. Waterhouse et al. “Jalview Version 2--a multiple sequence alignment editor and analysis workbench”. Bioinformatics. 25, 1189–1191 (2009).
I. C. Spyropoulos et al. “TMRPres2D: high quality visual representation of transmembrane protein models”. Bioinformatics. 20(17), 3258–3260 (2004).
A. Krogh et al. “Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes”. J. Mol. Biol. 19, 567–580 (2001).
R. Rezakhaniha et al. “Experimental investigation of collagen waviness and orientation in the arterial adventitia using confocal laser scanning microscopy”. Biomech. Model. Mechanobiol. 11, 461–473 (2012).
Acknowledgements
D.C. was supported by a Washington University School of Medicine Dean’s Fellowship. J.B.W. and C.E.S. were supported by a Ruth L. Kirschstein National Research Service Award from the Developmental Cardiology and Pulmonary Training Program (NIH T32 HL007873). P.Y.J. is an Established Investigator of the American Heart Association and the Lawrence J. & Florence A. DeGeorge Charitable Trust. This work was supported by the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital, the Children’s Heart Foundation, and the NIH (R01 HL105857).
Author information
Authors and Affiliations
Contributions
A.A.P., J.B.W., I.B.C. and P.Y.J. designed experiments. A.A.P., S.D.R., D.C., M.T.D., I.B.C., A.K.H., D.S., C.E.S., R.S.C. and P.Y.J. performed experiments. A.A.P., R.S.C., D.B.W. and P.Y.J. interpreted results. A.A.P. and P.Y.J. wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Panzer, A., Regmi, S., Cormier, D. et al. Nkx2-5 and Sarcospan genetically interact in the development of the muscular ventricular septum of the heart. Sci Rep 7, 46438 (2017). https://doi.org/10.1038/srep46438
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep46438
- Springer Nature Limited