Abstract
Hyperinvasive lineages of Neisseria meningitidis, which persist despite extensive horizontal genetic exchange, are a major cause of meningitis and septicaemia worldwide. Over the past 50 years one such lineage of meningococci, known as serogroup A, clonal complex 5 (A:cc5), has caused three successive pandemics, including epidemics in sub-Saharan Africa. Although the principal antigens that invoke effective immunity have remained unchanged, distinct A:cc5 epidemic clones have nevertheless emerged. An analysis of whole genome sequence diversity among 153 A:cc5 isolates identified eleven genetic introgression events in the emergence of the epidemic clones, which primarily involved variants of core genes encoding metabolic processes. The acquired DNA was identical to that found over many years in other, unrelated, hyperinvasive meningococci, suggesting that the epidemic clones emerged by acquisition of pre-existing metabolic gene variants, rather than ‘virulence’ associated or antigen-encoding genes. This is consistent with mathematical models which predict the association of transmission fitness with the emergence and maintenance of virulence in recombining commensal organisms.
Similar content being viewed by others
Introduction
Although N. meningitidis gives rise to 1.2 million cases of meningitis and severe sepsis disease each year1, asymptomatic colonisation of the human oropharynx is common, with population carriage rates of 10–30%2. As invasive disease does not contribute to person-to-person transmission, the meningococcus is an example of an ‘accidental’ pathogen3. Carried meningococci are highly diverse at loci encoding both antigens and metabolic functions, with much of this diversity generated by genetic reassortment, mediated by horizontal genetic transfer (HGT). Despite this diversity, meningococcal populations are highly structured into distinct genealogical groups or lineages, which are recognised by multilocus sequence typing (MLST) as ‘clonal complexes’ (ccs), which comprise closely related sequence types (STs)4. Epidemic meningococcal disease is caused by a subset of ccs, the ‘hyperinvasive lineages’, which persist for decades and during geographical spread5,6. Notwithstanding their propensity to cause invasive disease, hyperinvasive meningococci must also be efficient at asymptomatic transmission and several theoretical frameworks have been proposed to explain their emergence and persistence, including ‘strain structure theory’ which posits that the invasive phenotype can be stable in highly transmissible lineages3.
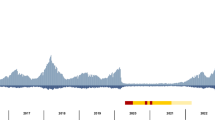
Since 1905, many of the largest recorded epidemics of meningococcal disease have occurred in the sub-Saharan African Meningitis Belt, mostly caused by serogroup A meningococci belonging to cc1, cc4, and cc5 (A:cc1; A:cc4; and A:cc5; Figure S1)7. These meningococci have also been responsible for a number of pandemics throughout the 20th century8, with A:cc1 meningococci dominant in Africa until they were replaced by A:cc5 meningococci during the Hajj-associated epidemics of 1987, part of the third A:cc5 global pandemic. Since that time, A:cc5 organisms have caused successive epidemics in the meningitis belt8 (Fig. 1A), including a large outbreak in 1997–1998, with more than 250,000 cases9. A:cc5 epidemic across the belt in the early 2000s coincided with the emergence of the novel cc5 genotypes A:cc5:ST-7 and A:cc5:ST-2859, which were identical to the original genotype (A:cc5:ST-5) at their principal antigens10,11,12. These disease outbreaks in the African Meningitis Belt prompted the development and implementation of the serogroup A conjugate vaccine PsA-TT (MenAfriVacTM)13, a polysaccharide–tetanus toxoid conjugate vaccine which targets the polysaccharide capsule of serogroup A meningococci.
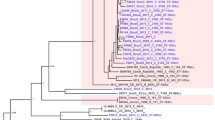
(A) Global spread of the ST-5 complex in three successive pandemics (red: first pandemic wave; blue: second pandemic wave; green: third pandemic wave; yellow: ST-2859). (B) Allele-based phylogenetic network of 153 whole genomes of the ST-5 complex. (C) Pie-charts of allelic diversity of six major sub-capsular antigens across the 153 isolates. Most isolates had a highly consistent antigenic profile, with a single dominant allele found for each antigen (PorA: 97.4% = allele P1.20,9; FetA: 97.4% = allele F3-1; PorB: 92.1% = allele 3–47; NadA: 99.3% = allele 7; OpcA: 96.1% = allele 3; fHbp: 83.1% = allele 39). PorA, FetA, PorB, NadA and fHbp have all been shown to induce an immune response and deployed in various protein-based vaccines15,16. See also Table S7. The maps were created using mapchart.net (www.mapchart.net).
The reasons for repeated pandemics and epidemics of antigenically highly uniform but distinct serogroup A meningococci remain poorly understood. Such marked levels of genetic and antigenic uniformity are unusual among meningococci, which exhibit high levels of genetic and antigenic diversity14 and the global spread of A:cc5 meningococcal variants provides an opportunity to study the emergence of new pandemic lineages with limited genetic variation, in an organism which is usually highly diverse as a consequence of extensive HGT. Understanding this stability has important implications for the continued use of the PsA-TT vaccine, the success of which depends on the antigenic stability of hyperinvasive serogroup A meningococci13. Using a gene-by-gene whole genome MLST (wgMLST) approach, we analysed 153 sets of WGS data from A:cc5 meningococci representative of the various A:cc5 pandemics and epidemics, to elucidate the diversification of this lineage over time and to investigate possible genetic factors driving the evolution of distinct variants.
Results and Discussion
We refer to the three A:cc5 pandemics and the outbreaks caused by A:cc5:ST-2859 as four separate “epidemic waves”. There was allelic variation at 50.1% (999/1993) of loci across the 153 meningococcal genomes analysed, but within epidemic waves the genomes were highly uniform, with an average of 67.2% of loci exhibiting identical alleles within each wave, and 87.2% of variants exhibiting alleles which were identical across >90% of all alleles within each wave. Eight major immunogenic sub-capsular antigens, many of which have been used in licensed protein-based meningococcal vaccines15,16, were highly conserved, with >99% of the A:cc5 meningococci exhibiting identical alleles (Fig. 1C; Table S7). Previous studies identified differences in six antigens (transferrin-binding protein B, IgA1 protease, OpaB, OpaD, FetA) and the lgt gene (involved in lipooligopolysaccharide synthesis) between isolates of the second and third pandemics8,17,18,19, and differences in Maf adhesins and pilin glycosylation loci among ST-7 and ST-2859 isolates20. The fact that the A:cc5 meningococci isolates manifest identical alleles at loci encoding six major antigens, which are known to elicit protective immune responses as assessed by serum bactericidal antibodies21, and which are also known to vary among meningococci to avoid protective immunity in Africa and elsewhere22, suggests that the evasion of host immune responses is unlikely to have played a major role in the emergence of the epidemic waves in Africa.
Although the genes encoding the major immunogenic antigens were highly conserved, a number of allelic differences were observed among the epidemic waves: there were 39 loci with alleles specific to the first pandemic; 69 loci with alleles specific to the second pandemic; and 95 loci with alleles specific to the third pandemic (Tables S2–4). A total of 73 loci were specific to ST-2859 genomes, with 14 loci absent compared to ST-5 isolates (Fig. 1B; Tables S5–6). The variable loci were annotated as encoding: metabolic functions (40.0%); genetic information processing (16.2%); environmental information processing (12.3%); other functions (6.6%); antigenic genes (3.7%); and cellular processes (2.2%); and there was no characterised function for 18.3% of these loci (Fig. 2A). The variable loci were dispersed around the chromosome, but when plotted consecutively against their position in a reference genome, allelic changes at several contiguous loci were observed (Fig. 2B), suggesting the introgression of large genomic regions (of up to 16.2 kb) from other meningococci via HGT in the emergence of these strains.
(A) Functional characterisations of alleles specific to each epidemic wave. (B) Plots of successive allelic changes against their position in the reference genome for each epidemic wave, with an accompanying plot of these changes annotated on the circular chromosome of Z2491 (or WUE2594 for ST-2859). Letters indicate areas of allelic changes which are adjacent on the chromosome. (C) Genomic areas of putative recombination between ST-2859 and ST-11/ST-167 strains. Comparison plots show hypothetical donor strains on the bottom level (either reference strain FAM18, an isolate of serogroup C ST-11, or M12 240332, a serogroup Y ST-167 complex strain), ST-2859 on the central level (isolate ERR052831), and reference strain WUE 2594 (serogroup A, ST-5 complex) on the top level (representative of the recipient strain). The arrows signify putative areas of recombination, and correspond to higher sequence identity shared between FAM18/M12 240332 and ST-2859, than the ancestral WUE 2594 strain and ST2859.
These putatively introgressed loci were examined for possible sources of HGT by comparison to allelic variation recorded in the PubMLST.org/neisseria database. Exact nucleotide matches to recorded meningococcal isolates in the database were observed at eight contiguous groups of loci (areas A, C, D, E, F, G, H and I in ST-2859 isolates; areas G and H in second pandemic wave isolates; and area H in third pandemic wave isolates; see Table 1, Fig. 2C). Areas G and H from the second pandemic, as well as Area H in the third pandemic, had exact matches to large numbers of globally distributed isolates belonging to cc11 hyperinvasive meningococci23. The oldest matching cc11 isolates dated from 1964, several years in advance of the second A:cc5 pandemic, which was consistent with these introgressions originating in cc11. The regions of contiguous allelic changes within ST-2859 isolates contained exact matches to isolates belonging to multiple hyperinvasive clonal complexes. The alleles from seven of these regions were present in globally-distributed cc11 isolates and were universally present in a group of W:cc11 strains circulating in Burkina Faso and Niger in 2001 and 2002. These W:cc11 isolates therefore represent plausible relatives of the donor strains involved in the emergence of the A:cc5:ST-2859 strain in sub-Saharan Africa. This timescale is consistent with the phylogenetic analyses of Lamelas et al., who calculated that the ST-2859 lineage emerged in Burkina Faso in 200020. Area F of A:cc5:ST-2859 also matched isolates from another lineage typically expressing serogroup Y, Y:cc167, with the earliest allele identified in an isolate from 1940. These data are consistent with the epidemic A:cc5:ST-2859 strain arising from multiple introgression events involving several hyperinvasive meningococci in a short time, which accounts for the rapid accumulation of diversity and the long branch lengths in allele-based phylogenies (Fig. 1).
Importantly these introgression events did not involve the major antigens, which are very different in these other lineages, as would be expected in an immune selection model. In addition, most of the introgressed alleles were present in older isolates in the PubMLST database (Table 1), with the earliest dating from 1937, demonstrating that the introgressed alleles are long-lived, having circulated over periods of several decades at least. HGT occurs frequently among carried meningococci3,14; however, given that the hyperinvasive meningococci represent a small minority of the carried population in Africa24, as elsewhere, the acquisition of multiple contiguous identical alleles by one hyperinvasive meningococcus from another, appears to be highly unlikely and is suggestive of a selective process. It is well established that rare HGT events, between and within Neisseria species, can be amplified in meningococcal populations by factors such as antibiotic25 use and immunological pressure26, and the data presented here are consistent with selection for tracts of metabolic gene variants within the A:cc5 genome.
The majority of putatively introgressed genes were annotated as having metabolic functions (62.5%, 25 out of 40 loci; 50% of all 50 loci, including those with unknown functions) (Figure S2). Differences in metabolic genes can contribute to the emergence of epidemic strains in a number of ways. First, there is increasing support for the idea that metabolic genes play important roles in pathogenesis and virulence in both meningococci27 and other bacterial pathogens28. Such metabolic adaptation could allow meningococci to exploit alternative host resources in invasive disseminated infections29, for example, and differences have been shown in the expression of metabolic genes between meningococci adhering to lung epithelial cells and growing in blood27,30,31. It is also plausible that differences in metabolic efficiency lead to differences in transmissibility among strains, as strains assimilate metabolites at different rates. Indeed, a recent study27 showed a significantly higher in vitro growth rate among meningococcal strains from hyperinvasive lineages compared to those from carried lineages.
Analysis of the predicted metabolic functions of the introgressed genes indicated that the largest category was energy metabolism (30%) (Figure S3). Notably, introgressed area G from isolates of the second pandemic encoded several subunits from the Na+-translocating NADH-quinone reductase complex, which carries out key redox reactions of the electron transport pathway, and are predicted to interact in the same functional network (Figure S4). Although we are not aware of any experimental data for the functional significance of Na+-translocating NADH-quinone reductase complex in N. meningitidis, there are experimental data from other Gram Negative pathogens suggesting that genetic variants of the Na+-translocating NADH-quinone reductase complex have different affinities for sodium ions in different species, thus influencing rates of NADH oxidation and energy transduction32. It is plausible that differences in energy metabolism among strains from the same species could result in differences in transmission phenotype. Given the importance of rapid growth of meningococci in the blood stream in the development of IMD in individuals, these changes also have implications for virulent phenotypes. Several of the other allelic differences were in loci assigned to genetic information processing functions (16.2%). Further, the functions of the genes in the genetic information processing category pertain to DNA replication, transcription, translation and repair, and it is therefore plausible that allelic differences may influence growth and replication rates33,34.
A predominant paradigm for changes in the frequency of epidemic clones is that antigenic genes vary over time by diversifying immune selection, while metabolic genes exhibit stabilising selection for conservation of function; indeed, the emergence of virulent strains in pathogenic bacteria has been associated with the import of, or mutation within, antibiotic resistance genes, antigens or virulence factors20. An alternate paradigm, however, posits that antigenic genes can be stable over time in non-overlapping combinations, as a consequence of between-host competition35. Such non-overlapping patterns have been observed among the PorA, FetA and Opa antigens of N. meningitidis, with many identical epitope combinations maintained over several decades5,36,37. By contrast, metabolic genes should evolve over time as a result of intense ecological competition within the host, so as to gain a competitive advantage against strains inhabiting the same antigenic niche: a phenomenon referred to as metabolic shift38. Our findings are consistent with mathematical models of immunological and ecological competition in N. meningitidis and also Streptococcus pneumoniae, which assume that metabolic differences between bacterial strains can lead to small differences in transmission fitness3,38. Simulations have shown successive replacement over time by strains with increasing metabolic fitness but similar antigenic properties.
Invasive disease is caused by a minority of meningococcal genotypes, the hyperinvasive lineages, which show strong temporal and geographic stability over decades and during global spread. Here, we show that HGT events among diverse long-lived hyperinvasive lineages (e.g. introgression into A:cc5:ST-5 from C:cc11:ST-11) can lead to the emergence of new epidemic strains (i.e. A:cc5:ST-2859). Our observations suggest that introgression events from one hyperinvasive lineage to another contribute to increased transmission fitness and/or increased virulence leading to the replacement of the previous variant. The introgressed genes, which constitute the majority of genetic differences among epidemic waves, encode metabolic functions, with many of the introgressed alleles present in other hyperinvasive isolates dating back several decades before the emergence of A:cc5:ST-2859.
Although immune escape may have played a minor role, our findings suggest that the most important events in the emergence of the A:cc5:ST-7 and A:cc5:ST-2859 epidemics were the acquisition of metabolic gene variants which have affected complex phenotypes, especially transmission fitness and possibly virulence. Such effects have been postulated in other pathogens38 and have potential implications for understanding epidemic bacteria and their prevention by vaccination as they indicate that large scale introgression events that alter the relationship between metabolic and antigenic types occur as a consequence of HGT. In principle such events could be induced by immune selection imposed by mass immunisation against principle antigens and on-going disease surveillance with genomic analysis is required to guard against such an eventuality.
Methods
The sequence reads for all ST-5 complex meningococcal genomes available as of 17/10/13 were downloaded from the European Nucleotide Archive (http://www.ebi.ac.uk/). The genomes were sequenced at the Wellcome Trust Sanger Institute in Cambridge, UK and the Institute for Genomic Sciences in Maryland, US (Table S1)20. Reads were assembled using an automated pipeline based on the Velvet algorithm, version 1.2.01. Annotation was carried out using the “autotagger” feature of the Bacterial Isolates Genome Sequence Database (BIGSdb) software39, which scans deposited sequences against defined loci in an automated BLAST process. The whole genome sequence data were compared using the BIGSdb Genome Comparator tool, which is implemented on the PubMLST website (www.pubmlst.org). The coding sequences within the annotation were extracted and compared against the reference strain Z2491 (accession number AL157959) using default parameters. Through Genome Comparator, unique allele sequences were labelled consecutively, allowing the identification of shared and unique alleles between isolates. Loci with alleles specific to each pandemic wave were identified, and functionally characterised according to the KEGG Orthology (KO) groupings of the KEGG database (www.kegg.jp). Genes with uncharacterised functions which did not fall into a KO category were blasted in the PFAM database (www.pfam.sanger.ac.uk), and functionally characterised accordingly. Genes without significant hits in the PFAM search were designated with an “unknown” function. Genome Comparator was run, in addition, with the ST-5 strain WUE 2594 (accession number FR774048) as a reference, to identify alleles specific to the ST-2859 strains.
Allele sequences were blasted against the PubMLST database to find the appropriate NEIS number and allele, which were then scanned in the database for sequence matches in other meningococci. The PubMLST database represents a large repository of whole genome data, with over 3700 whole genomes at the time of writing, from a variety of clonal complexes spanning a 78 year period. The Neighbour Net networks in Figures 1 and S1 were created using SplitsTree v440. Annotated plots of the genome (Fig. 2B) were created using the programme CG view41. The Artemis Comparison Tool42, funded by the Wellcome Trust, was used to create Fig. 2C.
Additional Information
How to cite this article: Watkins, E. R. and Maiden, M. C. J. Metabolic shift in the emergence of hyperinvasive pandemic meningococcal lineages. Sci. Rep. 7, 41126; doi: 10.1038/srep41126 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Jafri, R. Z. et al. Global epidemiology of invasive meningococcal disease. Popul. Health Metr. 11, 17 (2013).
Cartwright, K. A. V., Stuart, J. M., Jones, D. M. & Noah, N. D. The Stonehouse Survey - Nasopharyngeal Carriage Of Meningococci And Neisseria Lactamica. Epidemiol. Infect. 99, 591–601 (1987).
Buckee, C. O. et al. Role of selection in the emergence of lineages and the evolution of virulence in Neisseria meningitidis . Proc. Natl. Acad. Sci. USA 105, 15082–15087 (2008).
Maiden, M. C. J. et al. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95, 3140–3145 (1998).
Watkins, E. R. & Maiden, M. C. J. Persistence of Hyperinvasive Meningococcal Strain Types during Global Spread as Recorded in the PubMLST Database. PLoS One 7, e45349 (2012).
Caugant, D. A. et al. Intercontinental spread of a genetically distinctive complex of clones of Neisseria meningitidis causing epidemic disease. Proc. Natl. Acad. Sci. USA 83, 4927–4931 (1986).
Greenwood, B. Meningococcal meningitis in Africa. Trans. R. Soc. Trop. Med. Hyg. 93, 341–353 (1999).
Olyhoek, T., Crowe, B. A. & Achtman, M. Clonal Population Structure Of Neisseria-Meningitidis Serogroup-A Isolated From Epidemics And Pandemics Between 1915 And 1983. Rev. Infect. Dis. 9, 665–692 (1987).
Roberts, L. An ill wind, bringing meningitis. Science. 320, 1710–1715 (2008).
Nicolas, P., Norheim, G., Garnotel, E., Djibo, S. & Caugant, D. A. Molecular epidemiology of Neisseria meningitidis isolated in the African Meningitis Belt between 1988 and 2003 shows dominance of sequence type 5 (ST-5) and ST-11 complexes. J. Clin. Microbiol. 43, 5129–5135 (2005).
Sié, A. et al. ST2859 serogroup A meningococcal meningitis outbreak in Nouna Health District, Burkina Faso: a prospective study. Trop. Med. Int. Heal. 13, 861–868 (2008).
Huber, C. A. et al. Lack of antigenic diversification of major outer membrane proteins during clonal waves of Neisseria meningitidis serogroup A colonization and disease. FEMS Immunol. Med. Microbiol. 67, 4–10 (2012).
Daugla, D. M. et al. Effect of a serogroup A meningococcal conjugate vaccine (PsA-TT) on serogroup A meningococcal meningitis and carriage in Chad: A community study. Lancet 383, 40–47 (2014).
Jolley, K. A. et al. Carried meningococci in the Czech Republic: a diverse recombining population. J. Clin. Microbiol. 38, 4492–4498 (2000).
Pizza, M. et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 287, 1816–1820 (2000).
Bjune, G. et al. Effect of outer membrane vesicle vaccine against group B meningococcal disease in Norway. Lancet 338, 1093–1096 (1991).
Zhu, P. et al. Fit genotypes and escape variants of subgroup III Neisseria meningitidis during three pandemics of epidemic meningitis. Proc. Natl. Acad. Sci. USA 98, 5234–5239 (2001).
Norheim, G. et al. Characterization of Neisseria meningitidis isolates from recent outbreaks in Ethiopia and comparison with those recovered during the epidemic of 1988 to 1989. J. Clin. Microbiol. 44, 861–871 (2006).
Achtman, M. et al. A comparison of the variable antigens expressed by clone IV-1 and subgroup III of Neisseria meningitidis serogroup A. J. Infect. Dis. 165, 53–68 (1992).
Lamelas, A. et al. Emergence of a new epidemic Neisseria meningitidis serogroup A Clone in the African meningitis belt: high-resolution picture of genomic changes that mediate immune evasion. MBio 5, e01974–14 (2014).
Frasch, C. E., Borrow, R. & Donnelly, J. Bactericidal antibody is the immunologic surrogate of protection against meningococcal disease. Vaccine 27, 112–116 (2009).
Gasparini, R. et al. How the Knowledge of Interactions between Meningococcus and the Human Immune System Has Been Used to Prepare Effective Neisseria meningitidis Vaccines. J. Immunol. Res. 189153 (2015).
Lucidarme, J. et al. Genomic resolution of an aggressive, widespread, diverse and expanding meningococcal serogroup B, C and W lineage. J. Infect. 71, 544–552 (2015).
Greenwood, B. Meningococcal carriage in the African meningitis belt. Trop. Med. Int. Heal. 18, 968–978 (2013).
Spratt, B. G. et al. Recruitment of a penicillin-binding protein gene from Neisseria flavescens during the emergence of penicillin resistance in Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 86, 8988–8992 (1989).
Linz, B., Schenker, M., Zhu, P. & Achtman, M. Frequent interspecific genetic exchange between commensal Neisseriae and Neisseria meningitidis . Mol. Microbiol. 36, 1049–1058 (2000).
Schoen, C., Kischkies, L., Elias, J. & Ampattu, B. J. Metabolism and virulence in Neisseria meningitidis . Front. Cell. Infect. Microbiol. 4 (2014).
Shelburne, S. A., Davenport, M. T., Keith, D. B. & Musser, J. M. The role of complex carbohydrate catabolism in the pathogenesis of invasive streptococci. Trends Microbiol. 16, 318–325 (2008).
Pagliarulo, C. et al. Regulation and differential expression of gdhA encoding NADP-specific glutamate dehydrogenase in Neisseria meningitidis clinical isolates. Mol. Microbiol. 51, 1757–1772 (2004).
Echenique-Rivera, H. et al. Transcriptome analysis of Neisseria meningitidis in human whole blood and mutagenesis studies identify virulence factors involved in blood survival. PLoS Pathog 7, e1002027 (2011).
Hey, A. Transcriptional profiling of Neisseria meningitidis interacting with human epithelial cells in a long-term in vitro colonization model. Inf. Imm. 81, 4149–4159 (2013).
Verkhovsky, M. I. & Bogachev, A. V. Sodium-translocating NADH:quinone oxidoreductase as a redox-driven ion pump. Biochim et Biophys Acta Bioenerg 1797, 738–746 (2010).
Pierlé, S. A., Hammac, G. K., Palmer, G. H. & Brayton, K. A. Transcriptional pathways associated with the slow growth phenotype of transformed Anaplasma marginale . BMC Genomics 14, 272–280 (2013).
Dastgheyb, S. S. & Otto, M. Staphylococcal adaptation to diverse physiologic niches: an overview of transcriptomic and phenotypic changes in different biological environments. Future Microbiol. 10, 1981–95 (2015).
Gupta, S., Ferguson, N. & Anderson, R. Chaos, persistence, and evolution of strain structure in antigenically diverse infectious agents. Science 280, 912–915 (1998).
Callaghan, M. J. et al. The effect of immune selection on the structure of the meningococcal Opa protein repertoire. Plos Pathog. 4, e1000020 (2008).
Buckee, C. O., Gupta, S., Kriz, P., Maiden, M. C. J. & Jolley, K. A. Long-term evolution of antigen repertoires among carried meningococci. Proc. R. Soc. B-Biological Sci. 277, 1635–1641 (2010).
Watkins, E. R. et al. Vaccination Drives Changes in Metabolic and Virulence Profiles of Streptococcus pneumoniae . PLoS Pathog. 11, e1005034 (2015).
Jolley, K. A. & Maiden, M. C. J. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11, 595 (2010).
Huson, D. H. & Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 (2006).
Grant, J. R. & Stothard, P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36, 181–184 (2008).
Carver, T. J. et al. ACT: The Artemis comparison tool. Bioinformatics 21, 3422–3423 (2005).
Acknowledgements
E.R. Watkins was funded by the Jackson Scholarship from Merton College at the University of Oxford and the European Research Council (grant agreement no. 268904 – DIVERSITY). M.C.J. Maiden is supported by the Wellcome Trust (grant 087622). Sequence data were generated by the Wellcome Trust Sanger Institute in Cambridge (UK) and the Institute for Genomic Sciences in Maryland (US)20. This work made use of the Neisseria pubMLST website (http://pubmlst.org/neisseria/) developed by Martin Maiden and Keith Jolley and sited at the University of Oxford. The development of this site has been funded by the Wellcome Trust.
Author information
Authors and Affiliations
Contributions
E.R. Watkins analysed the genomic data, wrote the manuscript and made the figures. M.C.J. Maiden wrote the manuscript and conceived the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Watkins, E., Maiden, M. Metabolic shift in the emergence of hyperinvasive pandemic meningococcal lineages. Sci Rep 7, 41126 (2017). https://doi.org/10.1038/srep41126
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep41126
- Springer Nature Limited
This article is cited by
-
Common and distinctive genomic features of Klebsiella pneumoniae thriving in the natural environment or in clinical settings
Scientific Reports (2022)
-
Neisseria meningitidis: using genomics to understand diversity, evolution and pathogenesis
Nature Reviews Microbiology (2020)
-
First report of meningococcal ciprofloxacin resistance in Greece due to invasive isolates of the sequence type ST-3129
European Journal of Clinical Microbiology & Infectious Diseases (2020)