Abstract
Deoxyinosine (dI) occurs in DNA either by oxidative deamination of a previously incorporated deoxyadenosine residue or by misincorporation of deoxyinosine triphosphate (dITP) from the nucleotide pool during replication. To exclude dITP from the pool, mammals possess specific hydrolysing enzymes, such as inosine triphosphatase (ITPA). Previous studies have shown that deficiency in ITPA results in cell growth suppression and DNA instability. To explore the mechanisms of these phenotypes, we analysed ITPA-deficient human and mouse cells. We found that both growth suppression and accumulation of single-strand breaks in nuclear DNA of ITPA-deficient cells depended on MLH1/PMS2. The cell growth suppression of ITPA-deficient cells also depended on p53, but not on MPG, ENDOV or MSH2. ITPA deficiency significantly increased the levels of p53 protein and p21 mRNA/protein, a well-known target of p53, in an MLH1-dependent manner. Furthermore, MLH1 may also contribute to cell growth arrest by increasing the basal level of p53 activity.
Similar content being viewed by others
Introduction
For all organisms, maintenance of the integrity of genomic DNA and its precise transmission from cell to cell and from parents to offspring is fundamental to life. DNA, however, is susceptible to damage from various reactive molecules. Some DNA damage induces cell death or genetic mutation, and causes various disorders in humans, such as aging, cancer and hereditary diseases1,2. Base moieties of nucleic acids, which define genetic information, also suffer various chemical modifications, such as oxidation, deamination, methylation or halogenation3,4,5,6 that result in the generation of abnormal bases. These modifications can occur because of endogenous factors, such as reactive oxygen or nitrogen species, or after exposure to exogenous factors, such as ionizing radiation, ultraviolet light or chemical agents3,4,5,6. Various enzymatic reactions also generate abnormal bases in nucleic acids7,8. Direct modification of normal bases already incorporated in DNA is one of two main pathways for the accumulation of abnormal bases in DNA. The second pathway is the incorporation of abnormal deoxynucleoside triphosphates from the nucleotide pool into newly synthesized DNA during its replication. To avoid deleterious effects of the abnormal nucleotides, cells are equipped with specific enzymes to hydrolyse the abnormal nucleoside triphosphates to the corresponding monophosphates. These enzymes are known as nucleotide pool sanitizing enzymes9,10,11.
Deoxyinosine (dI) is an abnormal nucleoside and has hypoxanthine as its base moiety. Hypoxanthine is generated by oxidative deamination of adenine, which occurs in the presence of nitrous acid12, or via catalysis by specific enzymes, such as adenosine deaminase or AMP deaminase. dITP can be generated by oxidative deamination of dATP, and incorporated into DNA10,13,14. In addition, hypoxanthine is a base moiety of inosine monophosphate (IMP), which is a normal intermediate metabolite in the de novo purine nucleotide metabolism pathway. Pang et al. reported a large increase of dI in DNA in strains of Escherichia coli, and Saccharomyces cerevisiae unable to convert IMP to AMP or GMP, and unable to hydrolyze dITP/ITP15, suggesting the existence of a pathway from IMP, a normal nucleotide, to dI in DNA.
Previous studies in mammalian cells have revealed that inosine triphosphatase (ITPA), encoded by the ITPA gene, hydrolyses inosine triphosphate (ITP) and dITP to IMP and dIMP with essentially the same efficiency16,17. Itpa knockout (KO) mice die before weaning with features of growth retardation and heart failure18. These results show that ITP and dITP are produced under physiological conditions in living cells, and that they induce vital dysfunction unless hydrolysed by ITPA. Furthermore, Itpa KO mouse embryos had increased levels of deoxyinosine/inosine in DNA/RNA, and primary mouse embryonic fibroblasts (MEFs) derived from Itpa KO embryos exhibited prolonged doubling time and increased chromosome abnormalities and accumulation of single-strand breaks (SSBs) in nuclear DNA compared with primary MEFs prepared from wild-type embryos19.
We have previously performed a screen for ITP-binding proteins20 and revealed that nucleoside diphosphate linked moiety X-type motif16 (NUDT16), encoded by NUDT16, also hydrolyses (deoxy)inosine triphosphate and (deoxy)inosine diphosphate to (deoxy)inosine monophosphate. Knockdown of NUDT16 in either HeLa MR cells or ITPA-deficient MEF cells causes cell cycle delay in S phase, reduced cell proliferation, and increased accumulation of SSBs in nuclear DNA, suggesting that NUDT16, along with ITPA, has an important biological function in mammals as a sanitizing enzyme against inosine nucleotides.
The human ITPA gene has a polymorphic variant, P32T, which has decreased enzymatic activity through three mechanisms: protein instability, decreased rate of catalysis, and improper mRNA splicing21,22,23. The P32T variant is associated with potentially severe adverse drug reactions towards the thiopurine drugs, azathioprine and 6-mercaptopurine24. Furthermore, the P32T variant is related to protection against adverse effects of Ribavirin treatment in patients with hepatitis C25,26,27,28.
It has been reported that dI generated in DNA can be excised by several DNA repair systems in prokaryotes and eukaryotes. 3-Methyl-adenine DNA glycosylase II (AlkA) in Escherichia coli recognizes N-alkylpurine adducts, deaminated purine adducts, and lipid peroxidation-induced purine adducts in DNA, and cleaves N-glycosylic bonds within them. AlkA excises the hypoxanthine base from dI in DNA, forms an apurine/apyrimidine site (AP site) and initiates a base excision repair (BER) reaction29,30,31. Mammalian N-methylpurine-DNA glycosylase (MPG), which is known to excise at least 17 structurally diverse modified bases from DNA32, also removes hypoxanthine from double-stranded DNA (dsDNA), thus initiating BER33,34. Endonuclease V, a product of the nfi gene of E. coli, recognizes dI in DNA and cleaves the DNA at the second phosphodiester bond 3′ to the dI, leaving a nick and initiates the alternative excision repair (AER) system35,36,37. It has been reported that human and mouse also have endonuclease V homologs38,39. Mi et al. have reported that human endonuclease V (hENDOV) recognizes and repairs dI-containing dsDNA, according to the following order of dI pairings: dI in single-stranded DNA > dG:dI > dT:dI > dA:dI > dC:dI38. On the other hand, two other groups independently reported that recombinant hENDOV prefers inosine-containing single-stranded RNA but not dI-containing dsDNA as its substrate40,41,42, so the function of hENDOV against dI in DNA is currently controversial.
The DNA mismatch repair system (MMR) plays an important role in maintaining genome stability by correcting both base-base mismatches and insertion/deletion mispairs generated during DNA replication43. In the process of MMR, the newly synthesized DNA strand is incised and a DNA fragment longer than 150 bases and containing the incorrect base is removed. Defects in MMR increase the spontaneous mutation rate. Moreover, it has been established that the MMR system is required for cell cycle arrest and/or programmed cell death in response to certain types of DNA damage44,45,46.
In the present study, to understand the mechanism that causes cell growth delay and SSBs in nuclear DNA in ITPA-deficient cells, we analysed ITPA-deficient human and mouse cells, and revealed that ITPA deficiency induces MLH1/PMS2- and p53-dependent growth arrest and DNA instability in mammalian cells.
Results
MLH1 is involved in cell growth delay caused by NUDT16 knockdown in HeLa MR cells
We previously reported that knockdown of NUDT16 in HeLa MR cells, which are derived from human cervical cancer cells, caused growth delay20. We performed triple knockdown of NUDT16, MPG and ENDOV in HeLa MR cells to confirm whether these repair enzymes (MPG and ENDOV) are involved in the cell growth delay induced by knockdown of NUDT16 because bacterial endonuclease V was reported to cause DNA instability if dITP accumulates in bacterial cells13. Although single knockdown of MPG or ENDOV did not affect cell growth of HeLa MR cells, double knockdown of MPG and ENDOV induced significant cell growth delay similar to that seen for single knockdown of NUDT16 (Supplementary Figures S1 and S2). Knockdown of MPG and ENDOV in addition to NUDT16 knockdown (KD) did not rescue the phenotype of NUDT16 deficiency but caused additional cell growth delay. From these results, we suspected that other DNA repair enzymes might be involved in the growth delay caused by NUDT16 KD. We considered MLH1 as such a candidate because dI has been reported to pair with any normal deoxynucleotide in DNA, although its stability depends on opposite bases47. Double knockdown of MLH1 and NUDT16 rescued the cell growth delay induced by single knockdown of NUDT16. We also analysed the influence of ITPA KD; however, this did not affect HeLa MR cell growth (Supplementary Figure S1). These results suggest that the MMR pathway, but not MPG or ENDOV, is involved in the cell growth delay induced by accumulation of dITP or ITP caused by knockdown of NUDT16.
ITPA deficiency induces cell growth delay in normal human cells
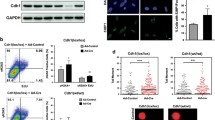
We previously reported that ITPA-deficient primary MEFs show growth delay compared with wild-type MEFs19. It is known that p53 and Rb proteins are inactivated by E6 and E7 gene products produced by human papilloma virus genes integrated in the HeLa genome48,49. As a result, the DNA damage response is not normal in HeLa cells. Therefore, we next used WI38 fibroblast cells, derived from human normal embryonic lung, to analyse ITPA deficiency in normal human cells. We analysed the ITPA cDNA sequence from WI38 cells and confirmed that WI38 expresses mRNA encoding wild-type ITPA protein (accession no. NP_258412.1). Western blot analysis showed that the level of ITPA in WI38 cells was essentially the same as in various other cell lines, including HeLa MR cells, another normal fibroblast cell, IMR90, and cell lines derived from human colorectal cancer (HCT116 and H414). The one exception was GM01617 (P32T) cells, a human fibroblast cell line that encodes the unstable ITPA P32T variant from both alleles (Fig. 1A)50. The level of NUDT16 protein in WI38 cells was also similar to that in HeLa MR cells and higher than that in human colorectal cancer cell lines (Fig. 1B). We performed knockdown experiments with the WI38 cell line, and transfection of siRNA for ITPA or NUDT16 efficiently decreased the expression of both genes (Fig. 1C). After transfection of control, ITPA or NUDT16 siRNA, proliferation of WI38 cells was analysed using a colorimetric assay. ITPA KD, but not NUDT16 KD, induced significant cell growth delay compared with control (Fig. 1D, upper panel). Whole cell extracts prepared from WI38 cells 4 days after siRNA transfection were subjected to western blot analysis of ITPA and NUDT16 proteins. This confirmed the efficient suppression of ITPA and/or NUDT16 expression (Fig. 1D, lower panels). These results reveal that ITPA deficiency induces cell growth delay in normal human cells as well as in MEF cells.
Knockdown of ITPA induces cell growth delay in WI38 cells.
(A) Western blot analysis of ITPA in human cell lines. Whole cell lysates were analysed with anti-ITPA and anti-GAPDH antibodies. The primary antibodies were detected with fluorophore-conjugated secondary antibodies. ITPA (green) and GAPDH (red) signals were detected and quantified using the Odyssey system. The bar graph shows relative levels of ITPA normalized to those of GAPDH. (B) Western blot analysis of NUDT16 in human cell lines. NUDT16 (green) and GAPDH (red) were detected in whole cell extracts as described for panel a. Normalized NUDT16 levels are shown in the bar graph. (C) Levels of ITPA and NUDT16 mRNAs in WI38 cells transfected with control, ITPA and/or NUDT16 siRNA. Two days after siRNA transfection, cells were subjected to analysis for RNA levels by real time quantitative RT-PCR. Data for each mRNA were normalized relative to that for 18S rRNA. Levels relative to control cells transfected with control siRNA are shown as the mean ± SD (n = 3). (D) Viability of WI38 cells transfected with control, ITPA and/or NUDT16 siRNA. Cell viability was measured using the Cell Counting Kit-8, 2, 3, 4, and 5 days after transfection. ITPA and NUDT16 protein levels 3 days after the siRNA transfections were confirmed by western blot analysis with antibodies against ITPA, NUDT16 and GAPDH. Relative protein levels normalized to GAPDH are shown under the blot images. Results are tested with two-way ANOVA, P = 0.0002; Tukey’s HSD post hoc test, P = 0.0256 (control versus ITPA siRNA), P = 0.0179 (control versus ITPA and NUDT16 siRNA). Data are presented as the mean ± SD (n = 3).
Cell growth delay caused by ITPA deficiency depends on MLH1
To confirm more precisely the contribution of MLH1 to the cell growth delay induced by ITPA deficiency, we performed ITPA KD in MLH1-deficient HCT116 cells, which are derived from human colorectal cancer cells, and MLH1-proficient H414 cells, which are derived from HCT116 cells as described in Supplementary Information (Figure S3). The HCT116 cell line is homozygous for a point mutation that changes serine (TCA) to stop (TAA) at amino acid residue 252 in exon 9 of the MLH1 gene located on chromosome 3, and thus this cell line is completely deficient in MLH1 protein51. HCT116 has wild-type p53, a near diploid karyotype (n = 45) and stable chromosome number52. HCT116 also lacks normal MSH3 expression53. We have established a cell line expressing wild-type MLH1 protein by targeted knock-in of the wild-type MLH1 coding sequence into one allele of the parental HCT116 cell line and named the line H414.
We transfected siRNAs for various genes into HCT116 and H414 cells and analysed their cell growth rates. Knockdown of ITPA induced significant cell growth delay in H414 cells. Triple knockdown of ITPA, MPG and ENDOV also caused growth delay in H414 cells, but the delay was essentially the same extent as that caused by knockdown of ITPA alone (Fig. 2A and S4A). Double knockdown of ITPA and MLH1 did not induce any change in cell growth, suggesting that MLH1 knockdown completely cancelled the cell growth delay induced by single knockdown of ITPA (Fig. 2B and S4B). On the other hand, in HCT116 cells, ITPA KD had no influence on their growth. Moreover, triple knockdown of ITPA, MPG and ENDOV had no effect on the proliferation of HCT116 cells (Fig. 2C and S4C). These results revealed that the cell growth delay caused by ITPA deficiency exclusively depends on MLH1 function, while MPG and ENDOV are not involved.
MLH1 is necessary for cell growth delay caused by ITPA knockdown in H414 cells.
(A) H414 cells were transfected with ITPA, MPG and/or ENDOV siRNA. Results are tested with two-way ANOVA, P < 0.0001; Tukey’s HSD post hoc test, P < 0.001 (control versus ITPA siRNA or control versus MPG/ENDOV/ITPA siRNA). (B) H414 cells were transfected with ITPA and/or MLH1 siRNA. Results are tested with two-way ANOVA, P < 0.0001; Tukey’s HSD post hoc test, P < 0.001 (control versus ITPA siRNA or MLH1 versus ITPA siRNA), P = 0.0014 (ITPA versus ITPA/MLH1 siRNA). (C) HCT116 cells were transfected with ITPA, MPG and/or ENDOV siRNA. Results tested with two-way ANOVA showed no significance. Cell viability was analysed using the Cell Counting Kit-8. Data are presented as the mean ± SD (n = 3).
Nuclear DNA instability induced by ITPA deficiency also depends on MLH1
We previously reported that Itpa-deficient primary MEFs accumulate SSBs in nuclear DNA and exhibit growth delay19. Therefore, we performed comet assays with HCT116 and H414 cells after ITPA KD to analyse the accumulation of SSBs in nuclear DNA. In alkaline conditions, H414 cells transfected with ITPA siRNA showed significantly increased comet tails and tail moments compared with cells transfected with control siRNA (Fig. 3A). In neutral conditions, there was no significant difference in comet tail moments between the two groups of cells (Fig. 3B). These results indicate that ITPA deficiency induced accumulation of SSBs or alkaline-labile sites such as abasic sites which are converted to SSBs in alkaline conditions, in the nuclear DNA of H414 cells. On the other hand, HCT116 cells transfected with ITPA siRNA did not exhibit any significant increase in comet tail moments in alkaline or neutral conditions compared with control HCT116 cells. These results suggest that ITPA deficiency induces accumulation of SSBs or alkaline-labile sites in nuclear DNA through an MLH1-dependent pathway. To analyze nuclear genome instability using a different assay system, we performed an in situ nick translation assay at neutral pH with H414 cells three days after siRNA transfection as described in Supplemental Materials and Methods, because the alkaline comet assay may detect alkaline-labile sites such as abasic sites. H414 cells were transfected independently three times for each siRNA. The cells with nick-dependent signals were significantly increased by ITPA KD from 35.9% in control to 62.0%, as shown in Fig. 3C. This result suggests that the increased tail moment following ITPA KD in Fig. 3A results from accumulated SSBs in nuclear genomic DNA.
MLH1 deficiency reduces accumulation of single-strand breaks in nuclear DNA caused by ITPA knockdown in H414 cells.
Cells were analysed using the comet assay 4 days after transfection with siRNAs. (A) Comet assay under alkaline conditions. Tail moments of at least 30 cells were calculated for each group. Comet images and calculated tail moments are shown in panels and box-and-whisker plots, respectively. Results tested with the Steel-Dwass test, P < 0.001 (control siRNA in H414 versus ITPA siRNA in H414), P = 0.0255 (control in HCT116 siRNA versus ITPA siRNA in H414), P = 0.0011 (ITPA siRNA in HCT116 versus ITPA siRNA in H414). (B) Comet assay under neutral conditions. Tail moments were not significantly increased in either group. The assays were independently performed three times. (C) In situ nick translation. The numbers of cells with or without fluorescein signal indicating nicks were counted. The mean ± SD (n = 3) of the ratio of nick-positive cells in each group is shown. Data were tested with Student’s t-test, P = 0.0169.
Deoxyinosine in the culture medium induces cell growth delay in H414 cells
The data described above indicated that the dITP accumulated in ITPA-deficient cells might be incorporated into nuclear DNA during its replication, and that SSBs in nuclear DNA were produced during the process of MMR, resulting in cell growth delay. To confirm whether intracellular dITP can cause cell growth delay, we analysed the effects of dI added into the culture medium. It is known that nucleosides in culture media can diffuse or be efficiently imported into cells by nucleoside transport proteins and can then be phosphorylated to nucleoside triphosphate by several kinases54,55,56. Addition of dI into the culture medium induced growth delay in H414 cells transfected with control siRNA in a dose-dependent manner (Fig. 4A), while H414 cells transfected with ITPA siRNA showed more severe delay in growth after addition of dI (Fig. 4B). On the other hand, addition of inosine did not change the proliferation rate of H414 cells transfected with control or ITPA siRNA (Fig. 4C,D). In HCT116 cells, there was no change in their proliferation rate after dI addition (Fig. 4E,F). These results suggest that dITP but not ITP induces cell growth delay in an MLH1-dependent manner. To clarify whether deoxyinosine nucleotides increase the single strand breakage of DNA, we subjected H414 ITPA-KD cells cultured with 250 μM dI to the comet assay (Fig. 4G). Overall comet tail moments were not significantly different between the dI-treated cells (dI+) and the control cells (dI−) after ITPA KD. However, 25th percentile values were 0.00215 in the control group and 0.45612 in the dI-treated group. These values are 0.29% and 38.12% of the 50th percentile value in each group respectively. Thus, dI treatment of ITPA-deficient cells tended to increase the ratio of cells with considerable tail moment. We cannot deny the possibility that supplementation of dI into the culture medium might disrupt nucleotide metabolism, thereby resulting in the incorporation of other nucleotide intermediates into the DNA or causing dNTP pool imbalances. Thus, we quantitatively measured dI content in genomic DNA of H414 cells after ITPA KD and dI supplementation of the medium (Fig. 4H). Two-way ANOVA analysis of the results revealed that each ITPA KD (P = 0.0096) and dI treatment (P = 0.0066) is an independent factor that significantly changes dI content in the DNA. Tukey’s HSD post hoc test among all groups showed that dI content is the highest in ITPA-KD/dI-treatment group with significant difference compared to those in ITPA KD without dI treatment (P = 0.0442) or control siRNA without dI treatment (P = 0.0048). These results indicate that the cellular responses induced by dI treatment, especially under ITPA deficiency, can be attributed to the incorporation of dI into the DNA, even if dI might disrupt nucleotide metabolism. Considering the contribution of dI incorporation into the DNA to its biological consequence, we noticed that majority of the basal dI content in H414 cells without ITPA KD or dI treatment may reflect dI generated by deamination of deoxyadenosine in the DNA during cell culture, because H414 cells without ITPA KD nor dI treatment have normal level of ITPA without dI supplementation, thereby avoiding dI incorporation into the DNA. On the other hand, the increase in dI content from the basal level after ITPA KD and/or dI treatment probably represent net incorporation of dI into DNA during DNA synthesis, and that the dI incorprated into the DNA, not the basal dI generated by deamination, may have caused growth arrest dependent on MLH1.
Deoxyinosine in the culture medium induces concentration-dependent cell growth delay in H414 cells.
Cells were transfected with siRNAs, and the culture medium was supplemented with deoxyinosine (dI) or inosine (rI) 24 h after transfection. Cell viability was measured using the Cell Counting Kit-8, 1, 2, 3, and 4 days after the supplementation. (A) H414 cells transfected with control siRNA were treated with the indicated concentration of deoxyinosine. Results tested with two-way ANOVA, P < 0.001; Tukey’s HSD post hoc test, P < 0.001, compared with control. (B) H414 cells transfected with ITPA siRNA were treated with dI. Results tested with two-way ANOVA, p < 0.001; Tukey’s HSD post hoc test, p < 0.001, compared with control. (C) H414 cells transfected with control siRNA were treated with rI. Results tested with two-way ANOVA shows no significance. (D) H414 cells transfected with ITPA siRNA were treated with rI. Results tested with two-way ANOVA shows no significance. (E) HCT116 cells transfected with control siRNA were treated with dI. Results tested with two-way ANOVA show no significance. (F) HCT116 cells transfected with ITPA siRNA were treated with dI. Results tested with two-way ANOVA show no significance. Data are presented as the mean ± SD (n = 3). (G) H414 cells cultured with 250 μM of dI for three days were analyzed by the comet assay under alkaline conditions. Tail moments of at least 18 cells were calculated for each group. The calculated tail moments are shown in box-and-whisker plots. The results were analysed with the Steel-Dwass test, and no significant increase in tail moments was observed between ITPA-KD cells with dI and ITPA-KD cells without dI. (H) DNA samples were prepared from H414 cells 4 days after 250 μM dI supplementaion, and their dI contents were analysed by LC–MS/MS. Results tested with two-way ANOVA, P = 0.0096 for ITPA-KD treatment, and P = 0.0066 for dI treatment. Tukey’s HSD post hoc test, P = 0.0048 (Control/none versus ITPA/dI), P = 0.0442 (ITPA/none versus ITPA/dI), and P = 0.0572 (Control/dI versus ITPA/dI). Data are presented as the mean ± SD (n = 3).
MLH1 deficiency partially rescues cell growth delay in ITPA-deficient primary MEFs but has no influence on the accumulation of deoxyinosine in nuclear DNA
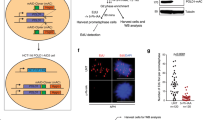
To examine ITPA deficiency-induced cell growth delay and its dependency on MLH1 in primary MEFs, we analysed proliferation rate of primary MEFs prepared from embryos derived from intercrosses of double heterozygous mice (Itpa+/−/Mlh1+/−), as described in the Supplementary Information. As in our previous report, Itpa-KO MEFs (Itpa−/−/Mlh1+/+) showed significant growth delay compared with wild-type MEFs (Itpa+/+/Mlh1+/+)19. Double KO MEFs (Itpa−/−/Mlh1−/−) also showed a delay in growth; however, the delay was significantly less than that in Itpa-single-KO MEFs (Itpa−/−/Mlh1+/+) (Fig. 5A). These results indicate that the growth delay in ITPA-deficient primary MEFs also depends, in part, on MLH1.
MLH1 deficiency partially rescues cell growth delay in ITPA-deficient primary mouse embryonic fibroblast (pMEF) cells, but had no effect on the accumulation of deoxyinosine in nuclear DNA of mouse embryos.
(A) Viability of pMEF cells. For each genotype, pMEF cells were independently prepared from at least five or six E14.5 embryos obtained from intercrossing double heterozygous mice (Itpa+/−/Mlh1+/−). Cell viability was measured using the Cell Counting Kit-8, 0, 2, 4 or 6 days after reseeding. Results tested with two-way ANOVA, P < 0.0001; Tukey’s HSD post hoc test, P < 0.0001 (Itpa+/+/Mlh1+/+ versus Itpa−/−/Mlh1+/+, Itpa+/+/Mlh1−/− versus Itpa−/−/Mlh1+/+), P =0.036 (Itpa−/−/Mlh1+/+ versus Itpa−/−/Mlh1−/−). (B) Deoxyinosine levels in nuclear DNA of mouse embryos. DNA samples were independently prepared from the heads of five E14.5 embryos for each genotype, and their dI contents were analysed by LC–MS/MS. Results tested with one-way ANOVA, P < 0.001. Tukey’s HSD post hoc test, P < 0.001 (Itpa+/+/Mlh1+/+ or Itpa+/+/Mlh1−/− versus Itpa−/−/Mlh1+/+ or Itpa−/−/Mlh1−/−). Data are presented as the mean ± SD (n = 5 or 6).
Next, we examined the accumulation of dI residues in nuclear DNA prepared from ITPA-and/or MLH1-deficient mouse embryos by liquid chromatography coupled with tandem mass spectrometry (LC–MS/MS). These analyses revealed that Itpa-KO embryos (Itpa−/−/Mlh1+/+) contained a significantly increased amount of dI in their nuclear DNA compared with wild-type embryos (Itpa+/+/Mlh1+/+). Furthermore, double KO embryos (Itpa−/−/Mlh1−/−) contained an increased amount of dI, similar to Itpa−/− and Mlh1+/+ embryos (Fig. 5B). In addition, we analysed the genotype of 74 mice obtained from intercrossing double heterozygous mice (Itpa+/−/Mlh1+/−). As shown in Supplementary Table S1, no Itpa−/− mice were detected among these offspring despite Mendel’s law predicting the identification of approximately 18 ITPA-deficient mice. From these data, we concluded that MLH1 deficiency can rescue the growth delay of ITPA-deficient mouse cells, but cannot rescue the perinatal lethality of ITPA-deficient mice.
ITPA deficiency induces G1 arrest in H414 cells
We analyzed the cell cycle of H414 and HCT116 cells 3 days after ITPA KD by flow cytometric analysis. ITPA KD significantly decreased the number of cells in S phase and increased the number of cells in G1 phase compared with negative control cells in H414 but not HCT116 cells (Fig. 6A), indicating G1 cell cycle arrest. To compare ITPA KD with other agents that induce cell cycle arrest, we treated H414 cells with N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) or fluorouracil (5-FU) and analyzed their cell cycles (Fig. 6B,C). MNNG induces cell cycle arrest through the MMR-mediated futile cycle57. The treatment of H414 cells with MNNG for 1 h induced accumulation of cells in G2 phase 72 h after the treatment in a dose-dependent manner, and treatment with 5-FU for 48 h caused S phase arrest. Thus, G1 arrest induced by ITPA deficiency is different from G2 or S phase cell cycle arrest induced by MNNG or 5-FU.
ITPA deficiency but not 5-FU or MNNG induced G1 cell cycle arrest.
Cell cycle analysis was performed using flowcytometry and ModFitLT software. (A) H414 and HCT116 cells were harvested three days after ITPA KD (n = 4). (B) H414 cells were treated with MNNG for 1 h, and then cultured without MNNG for 3 more days before harvest (n = 3). (C) H414 cells were cultured in the presence of 5-FU for 24 h and then harvested (n = 3). The mean ± SD is shown for G1, S, and G2 phases for each group. Results were tested with Pearson’s chi-square test or Fisher’s exact test in (A), P < 0.001 (total H414 cells with ITPA siRNA versus others, H414 cells with ITPA siRNA versus others in G1, and H414 cells with ITPA siRNA versus others in S phase).
PMS2 is necessary for cell growth suppression induced by ITPA deficiency, but MSH2 is dispensable
To evaluate contributions of other components of the MMR system, we examined effects of PMS2 or MSH2 deficiency in H414 cells. PMS2 forms a heterodimer with MLH1, and MSH2, together with MSH6 or MSH3, is essential for recognition of base mismatches in DNA. First, we performed knockdown of ITPA and PMS2 in H414 cells and analyzed cell growth and nuclear DNA damage with a Cell Counting Kit-8 and a comet assay respectively. Knockdown of PMS2 nullified both growth suppression and DNA damage in ITPA-deficient H414 cells (Fig. 7A,B). For MSH2, we established three MSH2-deficient H414 clones (#3–52, #3–113, and #3–119) using CRISPR/Cas9 technology. Each of the clones has a base insertion and/or a base deletion mutation on both MSH2 alleles as summarized in Supplementary Table S2. The mutations caused frame shifts and termination of translation by a new stop codon in exon 4 of each MSH2 allele. Western blot analysis confirmed MSH2 deficiency in these cell lines (Supplementary Figure S5A). Among the three clones, #3–113 proved to be resistant to 6-thioguanine, similar to MLH1-deficient HCT116 cells, thus clone #3–113 was deficient in MSH2 function (Supplementary Figure S5B). Each of these MSH2-deficient H414 clones showed significant suppression of cell proliferation after ITPA KD, similar to that in H414 cells with wild-type MSH2, indicating that the growth suppression caused by ITPA KD is independent of MSH2 (Fig. 7C).
PMS2, but not MSH2, contributes to cell growth arrest and DNA instability caused by ITPA knockdown.
H414 cells were transfected with ITPA and/or PMS2 siRNA, and then analysed using the Cell Counting Kit-8 (A) and comet assay in alkaline conditions (B) to assess cell viability and DNA damage, as in Figs 2 and 3A, respectively. For the comet assay, tail moments of at least 16 cells were calculated for each group. (C) Three H414-MSH2 deficient clones (#3–52 in upper, #3–113 in middle, and #3–119 in lower graph) were transfected with ITPA or negative control siRNA. Cell viability was analysed using the Cell Counting Kit-8. Data are presented as the mean ± SD (n = 3) in (A,C), or as box-and-whisker plots in (B). Results were tested with two-way ANOVA, P < 0.001; and Tukey’s HSD post hoc test (A), with two-way ANOVA, (control versus ITPA siRNA for each line) (C), or with the Steel-Dwass test, P < 0.0001 (control siRNA versus ITPA siRNA), P < 0.0001 (ITPA siRNA versus ITPA plus PMS2 siRNA) (B).
ITPA deficiency induces p53 and p21
To analyze the status of proteins related to cell cycle arrest, we harvested H414 cells 3 days after transfection of ITPA siRNA. Whole cell extracts were then subjected to western blot analysis. Levels of total p21 and p53 were increased after ITPA KD, but no induction of phosphorylation of CHK1, CHK2, or p38MAPK was observed (Fig. 8A). To analyze the induction of p53 and p21 proteins quantitatively, we repeated ITPA KD in H414 cells independently three times. Whole cell extracts were prepared 4 days later, and subjected to western blot analysis of p53 or p21 proteins (Fig. 8B). ITPA deficiency significantly increased both p53 and p21 protein levels. ITPA KD induced a mild increase of p53 protein phosphorylated at Ser15 or Ser33. To analyze p21 mRNA, H414 and HCT116 cells were harvested 3 days after transfection of ITPA siRNA, and extracted mRNA subjected to real time RT-PCR. The basal level of p21 mRNA in H414 cells was higher than that in HCT116 cells (p = 0.0226) (Fig. 8C). ITPA KD caused a significant increase in the level of p21 mRNA in H414 but not in HCT116 cells (p < 0.001). To address the contribution of p53, we performed p53 knockdown together with ITPA KD in H414 cells, and analyzed their cell cycle and cell proliferation using flow cytometry and the Cell Counting Kit-8 assay, respectively. ITPA KD alone caused a significant increase in the number of cells in G1 phase and a decrease in the number of cells in S phase, but did not cause any change in the cell cycle when p53 was also knocked down (Fig. 8D). p53 KD reduced the suppression of cell growth by ITPA KD although the effect was not significant (p = 0.1922) (Fig. 8E). p53 KD induced a reduction in the level of p21 mRNA, indicating that the elevated basal p21 mRNA level in H414 depends on p53 (Supplementary Figure S4D). ITPA KD did not increase p53 mRNA levels. Thus, the increase of p53 protein seems to depend on its stabilization.
ITPA knockdown up-regulates p53 protein and p21 mRNA/protein.
H414 and/or HCT116 cells were transfected with siRNAs. The cells were reseeded 24h after the transfection, and cultured further. (A) Three days after the transfection of control or ITPA siRNA, the H414 cells were analysed by western blotting with antibodies against indicated antigens. H414 cells treated with 20 μM etoposide for 8 h (ETP), or with 10 mM hydroxyurea for 2 h (HU) were used as positive control samples. (B) H414 cells were transfected with control or ITPA siRNA independently three times. After culture for 4 days, cells were harvested and whole cell extracts were subjected to western blot analysis with antibodies to p53 and p21. Blots were stained with Ponceau S to visualize total protein. The signals for p53, p21, and total protein are shown in the upper panels. The p53 or p21 levels normalized to Ponceau S staining are shown as the mean ± SD (n = 3) in the bar graphs of the lower panel. (C) Three days after the transfection of control or ITPA siRNA, HCT116 and H414 cells were harvested for an analysis of CDKN1A (p21) mRNA by real time RT-PCR. (D) Three days after the transfection of control, ITPA, and/or TP53 siRNA, the cell cycle of H414 cells was analysed. (E) At the indicated time points after the transfection of control, ITPA, and/or TP53 siRNA, H414 cells were analysed for cell viability. Results were tested with Student’s t-test, P = 0.0142 for p53, and P = 0.0133 for p21 (B), with two-way ANOVA and Tukey’s HSD post hoc test, P < 0.001 (H414 with control siRNA versus H414 with ITPA siRNA), P = 0.0226 (HCT116 with control siRNA versus H414 with control siRNA) (C,E) or with Fisher’s exact test, P = 0.0315 (H414 with control siRNA versus H414 with ITPA siRNA in G1), P = 0.0073 (H414 with control siRNA versus H414 with ITPA siRNA in S) (D).
Discussion
Last year, an early-infantile encephalopathy caused by ITPA mutations was reported58. The patients show encephalopathy, progressive microcephaly, seizures, variable cardiac defect, and early death. Most patients were small for the gestational age at birth. Recently, Nakauchi A et al. reported polymorphic variants of ITPA, including a P32T variant, as susceptibility genes for young-onset tuberculosis, which may result in immune system deficiency59. To develop treatments for these disorders it is crucial to understand the molecular mechanisms of the phenotypes caused by ITPA deficiency. ITPA deficiency may affect cells through multiple mechanisms, including competition between ATP and accumulated ITP, and alteration of RNA/DNA by incorporation of (d)ITP. In this paper, we focused on growth arrest and genome instability of proliferating cells with ITPA deficiency. Our results showed that cell growth arrest depends on p53 and MLH1 but not on ENDOV or MPG. However, this does not mean that ENDOV and MPG do not recognize dI in DNA. At first, we hypothesized that MLH1 contributes to the growth suppression of ITPA-deficient cells as a component of MMR. However, the growth arrest was independent of MSH2, an essential component of MMR, suggesting that MLH1 contributes to the growth suppression in an MMR-independent manner. ITPA KD induced SSBs in nuclear DNA and G1 phase cell cycle arrest in H414 cells, the mechanism of which may be different from the cell cycle arrest in G2 or S phase induced by MNNG or 5-FU. MLH1 is essential for both MMR and meiotic crossing-over in diploid germline cells. In the latter case, the MutS-γ complex, a heterodimer of MSH4 and MSH5, and the MutL-γ complex, a heterodimer of MLH1 and MLH3 play essential roles, however MSH2 complexes do not60. In addition, Siehler et al. reported MLH1-dependent and MSH2-independent suppression of homologous recombination by using MLH1- or MSH2-deficient human cell lines derived from colorectal adenocarcinoma61. Thus, our finding reveals another MMR-independent role of MLH1 in non-germline cells. The stabilization of p53 seems to play a central role in the ITPA deficiency-induced growth arrest. Previously, we observed S phase cell cycle arrest of HeLa MR cells after NUDT16 KD20. Although degradation of p53 and Rb proteins are enhanced by HPV18 E6 and E7 proteins in HeLa MR cells, some chemical treatments stabilize p53 in HeLa cells62. Such reversible interference might cause the difference between HeLa MR and H414 cells after NUDT16 or ITPA KD. Accumulated dITP may be recognized by DNA polymerases during DNA replication or DNA repair, and may stall the DNA polymerase complex. Repeated proofreading by the DNA polymerase is a candidate mechanism for polymerase stalling. Fujiwara H. et al. reported that Deep Vent polymerase (Exo−) but not Deep Vent (Exo+) could amplify DNA using dI-containing primers, suggesting that the proofreading function of some DNA polymerases may recognize a base pair of hypoxanthine and a normal base as a mismatch pair63. The stalled replication complexes or dI in DNA itself might induce SSBs in DNA dependent on MMR-independent function of the MLH1/PMS2 complex (Fig. 9) because our data indicate that MLH1 and PMS2 are required to induce SSBs in DNA (Figs 3A and 7B) and growth arrest (Figs 2B and 3A) under ITPA-deficient conditions. These findings indicate that the MLH1/PMS2 heterodimer functions as a DNA endonuclease in ITPA-deficient cells. Moreover, MLH1 is essential to keep higher basal p53 activity (Figs 8C and S4D) and induced levels of p53 protein and p21 mRNA/protein in H414 compared with HCT116 cells. Activation of p53 may depend on formation of SSBs in DNA, which may lead to undetectable levels of double-stand breaks, or depend on interaction between MLH1 and ATM as reported previously64,65. Induced p53 itself and/or its downstream effectors including p21 may cause G1 arrest. In our previous report using MLH1-proficient HCT116 + Chr3 and MLH1-deficient HCT116 cells, we observed higher p21 mRNA levels in MLH1-proficient cells66. Cordycepin-treatment induced higher p21 protein levels in HCT116 + Chr3 cells compared with MLH1-deficient HCT116 + Chr2 cells67. In addition, several groups reported MLH1-dependent activation of p5368,69,70. It seems that an MLH1-dependent higher basal level of p53 activity and MLH1-dependent activation of p53 by ITPA-deficiency synergistically promote cell growth arrest in the ITPA-deficient cells. To elucidate these precise mechanisms, further study is necessary.
Proposed mechanism of G1 cell cycle arrest induced by dITP.
In the absence of ITPA, accumulated dITP in the nucleotide pool is utilized as a substrate for DNA synthesis by DNA polymerase, thus stalling the polymerase. The stalled polymerase or deoxyinosine incorporated into DNA triggers MLH1/PMS2-dependent and MSH2-independent formation of SSBs in DNA. Accumulation of SSBs in DNA or other events caused by MLH1/PMS2 may induce G1 cell cycle arrest via stabilization of p53 protein. The p53 can induce its transcriptional targets including CDKN1A (p21) or affect other targets by direct interaction. MLH1 also increases the basal activity of p53.
Methods
Animal Experiments
All animals were maintained in an air-conditioned, light/time-controlled, specific-pathogen-free room. All studies were approved by the Animal Care and Use Committee of Kyushu University. All protocols were performed in compliance with the “Fundamental Guidelines for Proper Conduct of Animal Experiment and Related Activities in Academic Research Institutions under the jurisdiction of the Ministry of Education, Culture, Sports, Science and Technology of Japan”.
Oligonucleotides
Synthetic oligonucleotides are listed in Supplementary Table S3. All oligonucleotides were purchased from Applied Biosystems (Foster City, CA, USA), Integrated DNA technologies (Coralville, IA, USA) or Fasmac (Kanagawa, Japan).
Cell Culture
Human fibroblast GM01617 (P32T) cells were obtained from the Coriell Institute (Camden, NJ, USA). WI38 and IMR90 cells were obtained from Japan Cancer Research Resources Bank (Tokyo, Japan). HCT116 cells were obtained from ATCC (Manassas, VA). A Mer− variant of HeLaS3 strain, HeLa MR71, was kindly supplied by Dr. Rufus S. Day III, National Cancer Institute, NIH (Frederick, MD), and has been maintained in our laboratory. WI38, P32T, and IMR90 cells were routinely grown in minimum essential medium α (Life Technologies, Carlsbad, CA, USA). HeLa MR, HCT116 and H414 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies). Both media were supplemented with 10% heat-inactivated foetal bovine serum (FBS; PAA laboratories GmbH, Pasching, Austria), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Life Technologies). These cell lines were cultured at 37 °C in a 5% CO2 atmosphere.
Cell Proliferation Assay
Cell proliferation was analysed with a Cell Counting Kit-8, a colorimetric assay kit using WST-8 reagent (Dojindo, Kumamoto, Japan) according to the manufacturer’s instructions. This assay is similar to the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl-tetrazolium bromide (MTT) assay. Cells were seeded at a density of 500 cells per well in 96-well plates. After cultivation, WST-8 reagent was added to each well and cells were incubated for an additional 4 h. Absorbance of each sample was measured at 450 nm using an Infinite 200 Pro microplate reader (Tecan, Salzburg, Austria).
Cell Cycle Analysis
To analyze the cell cycle, flow cytometric analysis of isolated nuclei samples was performed as previously described20 using a FACSCalibur, CellQuest (BD Biosciences, San Jose, CA, USA) and ModFit LT Version 3 software (Verity Software House, Topsham, ME, USA).
Real-Time Quantitative RT-PCR
Total RNA from each cell culture was isolated with ISOGEN RNA extraction reagent (Nippon Gene, Tokyo, Japan). The relative levels of ITPA, NUDT16, MPG, ENDOV, MLH1, CDKN1A (p21), TP53 (p53) and PMS2 mRNA were determined by real-time quantitative RT-PCR, according to previously described methods20,72. Briefly, 20 μg of each total RNA was treated with 10 units of RNase-free DNase I (Roche Applied Science, Penzberg, Germany) at 37 °C for 1 h, and then purified by phenol/chloroform treatment and ethanol precipitation. cDNA was synthesized from 2 μg of DNA-free total RNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems) using random primers in a total volume of 20 μl. Real-time quantitative PCR was performed to measure the levels of each mRNA using a Thermal Cycler Dice® Real-Time System Single (Takara, Kyoto, Japan) with 10 ng cDNA, 200 nM primers and Thunderbird® SYBR® qPCR Mix (Toyobo, Osaka, Japan) in a total volume of 25 μl. The specificity of the PCR products was established by dissociating curve analysis. No primer dimers were observed. The mRNA level of each gene was normalized to that of 18S rRNA.
Comet Assay
Nuclear DNA fragmentation was monitored using a Comet Assay Kit and Comet Assay Electrophoresis System II (Trevigen, Gaithersburg, MD, USA) under alkaline and neutral conditions, according to the manufacturer’s instructions. H414 and HCT116 cells were transfected with control or ITPA siRNA. After incubation for 4 days, the cells were separately embedded in soft agarose on glass slides and then subjected to the assay. Comet images from more than 30 cells for each siRNA were captured using an Axioskop 2 plus equipped with an AxioCam (Carl Zeiss MicroImaging Japan, Tokyo, Japan), and were analysed using the Comet Assay Software Project (CASP) program73 to quantify tail moment.
Statistical Analysis
Statistical analyses were conducted using JMP 11.00 (SAS Institute, Cary, NC, USA). To assess statistical significance, we performed 1-way or 2-way analysis of variance (ANOVA); results obtained by standard least square fits are shown. For multiple comparison tests, ANOVA analyses were followed by a post hoc Tukey’s honestly significant difference (HSD) test. The Steel-Dwass test, Pearson’s chi-square test or Fisher’s exact test was applied for non-parametric multiple comparisons. The threshold P values for statistical significance were <0.05 (*), <0.01 (**), and <0.001 (***).
Additional Information
How to cite this article: Yoneshima, Y. et al. Deoxyinosine triphosphate induces MLH1/PMS2- and p53-dependent cell growth arrest and DNA instability in mammalian cells. Sci. Rep. 6, 32849; doi: 10.1038/srep32849 (2016).
References
Ames, B. & Gold, L. Endogenous Mutagens and the Causes of Aging and Cancer. Mutat Res 250, 3–16, doi: 10.1016/0027-5107(91)90157-j (1991).
Ames, B., Shigenaga, M. & Hagen, T. Oxidants, Antioxidants, and the Degenerative Diseases of Aging. Proc Natl Acad Sci USA 90, 7915–7922, doi: 10.1073/pnas.90.17.7915 (1993).
Dizdaroglu, M., Jaruga, P., Birincioglu, M. & Rodriguez, H. Free radical-induced damage to DNA: mechanisms and measurement. Free Radic Biol Med 32, 1102–1115 (2002).
Shrivastav, N., Li, D. & Essigmann, J. M. Chemical biology of mutagenesis and DNA repair: cellular responses to DNA alkylation. Carcinogenesis 31, 59–70, doi: 10.1093/carcin/bgp262 (2010).
Knutson, C. G. et al. Chemical and cytokine features of innate immunity characterize serum and tissue profiles in inflammatory bowel disease. Proc Natl Acad Sci USA 110, E2332–E2341, doi: 10.1073/pnas.1222669110 (2013).
Prestwich, E. G. et al. Increased levels of inosine in a mouse model of inflammation. Chem Res Toxicol 26, 538–546, doi: 10.1021/tx300473n (2013).
Jaszczur, M., Bertram, J. G., Pham, P., Scharff, M. D. & Goodman, M. F. AID and Apobec3G haphazard deamination and mutational diversity. Cel Mol Life Sci 70, 3089–3108, doi: 10.1007/s00018-012-1212-1 (2013).
Koito, A. & Ikeda, T. Intrinsic immunity against retrotransposons by APOBEC cytidine deaminases. Front Microbiol 4, 28, doi: 10.3389/fmicb.2013.00028 (2013).
Nakabeppu, Y., Oka, S., Sheng, Z., Tsuchimoto, D. & Sakumi, K. Programmed cell death triggered by nucleotide pool damage and its prevention by MutT homolog-1 (MTH1) with oxidized purine nucleoside triphosphatase. Mutat Res 703, 51–58, doi: 10.1016/j.mrgentox.2010.06.006 (2010).
Sakumi, K. et al. ITPA protein, an enzyme that eliminates deaminated purine nucleoside triphosphates in cells. Mutat Res 703, 43–50, doi: 10.1016/j.mrgentox.2010.06.009 (2010).
el-Hajj, H. H., Zhang, H. & Weiss, B. Lethality of a dut (deoxyuridine triphosphatase) mutation in Escherichia coli. J. Bacteriol. 170, 1069–1075 (1988).
Shapiro, R. & Pohl, S. H. The reaction of ribonucleosides with nitrous acid. Side products and kinetics. Biochemistry 7, 448–455 (1968).
Bradshaw, J. S. & Kuzminov, A. RdgB acts to avoid chromosome fragmentation in Escherichia coli. Mol Microbiol 48, 1711–1725 (2003).
Mangerich, A. et al. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc Natl Acad Sci USA 109, E1820–E1829, doi: 10.1073/pnas.1207829109 (2012).
Pang, B. et al. Defects in purine nucleotide metabolism lead to substantial incorporation of xanthine and hypoxanthine into DNA and RNA. Proc Natl Acad Sci USA 109, 2319–2324, doi: 10.1073/pnas.1118455109 (2012).
Behmanesh, M. et al. Characterization of the structure and expression of mouse Itpa gene and its related sequences in the mouse genome. DNA Res 12, 39–51, doi: 10.1093/dnares/12.1.39 (2005).
Lin, S. R. et al. Cloning, expression, and characterization of a human inosine triphosphate pyrophosphatase encoded by the ITPA gene. J Biol Chem 276, 18695–18701, doi: 10.1074/jbc.M011084200 (2001).
Behmanesh, M. et al. ITPase-deficient mice show growth retardation and die before weaning. Cell Death Differ 16, 1315–1322, doi: 10.1038/cdd.2009.53 (2009).
Abolhassani, N. et al. NUDT16 and ITPA play a dual protective role in maintaining chromosome stability and cell growth by eliminating dIDP/IDP and dITP/ITP from nucleotide pools in mammals. Nucleic Acids Res 38, 2891–2903, doi: 10.1093/nar/gkp1250 (2010).
Iyama, T., Abolhassani, N., Tsuchimoto, D., Nonaka, M. & Nakabeppu, Y. NUDT16 is a (deoxy)inosine diphosphatase, and its deficiency induces accumulation of single-strand breaks in nuclear DNA and growth arrest. Nucleic Acids Res 38, 4834–4843, doi: 10.1093/nar/gkq249 (2010).
Arenas, M., Duley, J., Sumi, S., Sanderson, J. & Marinaki, A. The ITPA c.94C > A and g.IVS2 + 21A > C sequence variants contribute to missplicing of the ITPA gene. Biochim Biophys Acta 1772, 96–102, doi: 10.1016/j.bbadis.2006.10.006 (2007).
Herting, G., Barber,K., Zappala, M. R., Cunningham, R. P. & Burgis, N. E. Quantitative in vitro and in vivo characterization of the human P32T mutant ITPase. Biochim Biophys Acta 1802, 269–274, doi: 10.1016/j.bbadis.2009.11.002 (2010).
Cao, H. N. & Hegele, R. A. DNA polymorphisms in ITPA including basis of inosine triphosphatase deficiency. J Hum Genet 47, 620–622, doi: 10.1007/s100380200095 (2002).
Sumi, S. et al. Genetic basis of inosine triphosphate pyrophosphohydrolase deficiency. Hum Genet 111, 360–367, doi: 10.1007/s00439-002-0798-z (2002).
Fellay, J. et al. ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature 464, 405–408, doi: 10.1038/nature08825 (2010).
Ochi, H. et al. ITPA Polymorphism Affects Ribavirin-Induced Anemia and Outcomes of Therapy-A Genome-Wide Study of Japanese HCV Virus Patients. Gastroenterology 139, 1190−+, doi: 10.1053/j.gastro.2010.06.071 (2010).
Sakamoto, N. et al. ITPA gene variant protects against anemia induced by pegylated interferon-alpha and ribavirin therapy for Japanese patients with chronic hepatitis C. Hepatol Res 40, 1063–1071, doi: 10.1111/j.1872-034X.2010.00741.x (2010).
Thompson, A. J. et al. Variants in the ITPA Gene Protect Against Ribavirin-Induced Hemolytic Anemia and Decrease the Need for Ribavirin Dose Reduction. Gastroenterology 139, 1181−+, doi: 10.1053/j.gastro.2010.06.016 (2010).
Cunningham, R. P. DNA glycosylases. Mutat Res 383, 189–196, doi: 10.1016/s0921-8777(97)00008-6 (1997).
Saparbaev, M. & Laval, J. Excision of Hypoxanthine from Dna Containing Dimp Residues by the Escherichia-Coli, Yeast, Rat, and Human Alkylpurine Dna Glycosylases. Proc Natl Acad Sci USA 91, 5873–5877, doi: 10.1073/pnas.91.13.5873 (1994).
Yamagata, Y. et al. Three-dimensional structure of a DNA repair enzyme, 3-methyladenine DNA glycosylase II, from Escherichia coli. Cell 86, 311–319 (1996).
Singer, B. & Hang, B. What structural features determine repair enzyme specificity and mechanism in chemically modified DNA? Chem Res Toxicol 10, 713–732, doi: 10.1021/tx970011e (1997).
Adhikari, S., Manthena, P. V., Ueren, A. & Roy, R. Expression, purification and characterization of codon-optimized human N-methylpurine-DNA glycosylase from Escherichia coli. Protein Expr Purif 58, 257–262, doi: 10.1016/j.pep.2007.12.001 (2008).
Saparbaev, M., Mani, J. C. & Laval, J. Interactions of the human, rat, Saccharomyces cerevisiae and Escherichia coli 3-methyladenine-DNA glycosylases with DNA containing dIMP residues. Nucleic Acids Res 28, 1332–1339, doi: 10.1093/nar/28.6.1332 (2000).
He, B., Qing, H. & Kow, Y. W. Deoxyxanthosine in DNA is repaired by Escherichia coli endonuclease V. Mutat Res 459, 109–114, doi: 10.1016/s0921-8777(99)00063-4 (2000).
Yao, M., Hatahet, Z., Melamede, R. & Kow, Y. Purification and Characterization of a Novel Deoxyinosine-Specific Enzyme, Deoxyinosine 3′ Endonuclease, from Escherichia-Coli. J Biol Chem 269, 16260–16268 (1994).
Yao, M. & Kow, Y. Strand-Specific Cleavage of Mismatch-Containing Dna by Deoxyinosine 3′-Endonuclease from Escherichia-Coli. J. Biol Chem 269, 31390–31396 (1994).
Mi, R., Alford-Zappala, M., Kow, Y. W., Cunningham, R. P. & Cao, W. Human endonuclease V as a repair enzyme for DNA deamination. Mutat Res 735, 12–18, doi: 10.1016/j.mrfmmm.2012.05.003 (2012).
Moe, A. et al. Incision at hypoxanthine residues in DNA by a mammalian homologue of the Escherichia coli antimutator enzyme endonuclease V. Nucleic Acids Res 31, 3893–3900, doi: 10.1093/nar/gkg472 (2003).
Fladeby, C. et al. The Human Homolog of Escherichia coli Endonuclease V Is a Nucleolar Protein with Affinity for Branched DNA Structures. PLoS One 7, e47466–e47466, doi: 10.1371/journal.pone.0047466 (2012).
Morita, Y. et al. Human endonuclease V is a ribonuclease specific for inosine-containing RNA. Nat Commun 4 (2013).
Vik, E. S. et al. Endonuclease V cleaves at inosines in RNA. Nat Commun 4 (2013).
Li, G. M. Mechanisms and functions of DNA mismatch repair. Cell Res 18, 85–98, doi: 10.1038/cr.2007.115 (2008).
Jiricny, J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol 7, 335–346, doi: 10.1038/nrm1907 (2006).
Kunkel, T. A. & Erie, D. A. DNA mismatch repair. Ann Rev Biochem 74, 681–710, doi: 10.1146/annurev.biochem.74.082803.133243 (2005).
Modrich, P. Mechanisms in eukaryotic mismatch repair. J Biol Chem 281, 30305–30309, doi: 10.1074/jbc.R600022200 (2006).
Martin, F., Castro, M., Aboulela, F. & Tinoco, I. Base-Pairing Involving Deoxyinosine - Implications for Probe Design. Nucleic Acids Res 13, 8927–8938, doi: 10.1093/nar/13.24.8927 (1985).
Chellappan, S. et al. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc Natl Acad Sci USA 89, 4549–4553 (1992).
Kessis, T. D. et al. Human papillomavirus 16 E6 expression disrupts the p53-mediated cellular response to DNA damage. Proc Natl Acad Sci USA 90, 3988–3992 (1993).
Stepchenkova, E. I. et al. Functional study of the P32T ITPA variant associated with drug sensitivity in humans. J Mol Biol 392, 602–613, doi: 10.1016/j.jmb.2009.07.051 (2009).
Boyer, J. C. et al. Microsatellite instability, mismatch repair deficiency, and genetic defects in human cancer cell lines. Cancer Res 55, 6063–6070 (1995).
Brattain, M. G., Fine, W. D., Khaled, F. M., Thompson, J. & Brattain, D. E. Heterogeneity of malignant cells from a human colonic carcinoma. Cancer Res 41, 1751–1756 (1981).
Haugen, A. C. et al. Genetic instability caused by loss of MutS homologue 3 in human colorectal cancer. Cancer Res 68, 8465–8472, doi: 10.1158/0008-5472.CAN-08-0002 (2008).
Bourdais, J. et al. Cellular phosphorylation of anti-HIV nucleosides. Role of nucleoside diphosphate kinase. J Biol Chem 271, 7887–7890 (1996).
Johansson, M., van Rompay, A. R., Degreve, B., Balzarini, J. & Karlsson, A. Cloning and characterization of the multisubstrate deoxyribonucleoside kinase of Drosophila melanogaster. J Biol Chem 274, 23814–23819 (1999).
Van Rompay, A. R., Johansson, M. & Karlsson, A. Substrate specificity and phosphorylation of antiviral and anticancer nucleoside analogues by human deoxyribonucleoside kinases and ribonucleoside kinases. Pharmacol & Therap 100, 119–139 (2003).
Fu, D., Calvo, J. A. & Samson, L. D. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer 12, 104–120, doi: 10.1038/nrc3185 (2012).
Kevelam, S. H. et al. Recessive ITPA mutations cause an early-infantile encephalopathy. Ann Neurol 78, 649–658, doi: 10.1002/ana.24496 (2015).
Nakauchi, A. et al. Identification of ITPA on chromosome 20 as a susceptibility gene for young-onset tuberculosis. Hum Genome Var 3, 15067, doi: 10.1038/hgv.2015.67 (2016).
Manhart, C. M. & Alani, E. Roles for mismatch repair family proteins in promoting meiotic crossing over. DNA Repair (Amst) 38, 84–93, doi: 10.1016/j.dnarep.2015.11.024 (2016).
Siehler, S. Y. et al. Human MutL-complexes monitor homologous recombination independently of mismatch repair. DNA Repair (Amst) 8, 242–252, doi: 10.1016/j.dnarep.2008.10.011 (2009).
Wesierska-Gadek, J., Schloffer, D., Kotala, V. & Horky, M. Escape of p53 protein from E6-mediated degradation in HeLa cells after cisplatin therapy. International journal of cancer. J Internat du Cancer 101, 128–136, doi: 10.1002/ijc.10580 (2002).
Fujiwara, H., Fujiwara, K. & Hashimoto, K. Pcr with Deoxyinosine-Containing Primers Using DNA-Polymerases with Proofreading Activity. Pcr Meth Appl 4, 239–240 (1995).
Brown, K. D. et al. The mismatch repair system is required for S-phase checkpoint activation. Nat Genet 33, 80–84, doi: 10.1038/ng1052 (2003).
Wang, Y. et al. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev 14, 927–939 (2000).
Oka, S., Leon, J., Tsuchimoto, D., Sakumi, K. & Nakabeppu, Y. MUTYH, an adenine DNA glycosylase, mediates p53 tumor suppression via PARP-dependent cell death. Oncogenesis 3, e121, doi: 10.1038/oncsis.2014.35 (2014).
Imesch, P., Goerens, A., Fink, D. & Fedier, A. MLH1-deficient HCT116 colon tumor cells exhibit resistance to the cytostatic and cytotoxic effect of the poly(A) polymerase inhibitor cordycepin (3′-deoxyadenosine) in vitro. Oncol Lett 3, 441–444, doi: 10.3892/ol.2011.504 (2012).
Duckett, D. R., Bronstein, S. M., Taya, Y. & Modrich, P. hMutSalpha- and hMutLalpha-dependent phosphorylation of p53 in response to DNA methylator damage. Proc Natl Acad Sci USA 96, 12384–12388 (1999).
Luo, Y., Lin, F. T. & Lin, W. C. ATM-mediated stabilization of hMutL DNA mismatch repair proteins augments p53 activation during DNA damage. Mol Cell Biol 24, 6430–6444, doi: 10.1128/MCB.24.14.6430-6444.2004 (2004).
Yanamadala, S. & Ljungman, M. Potential role of MLH1 in the induction of p53 and apoptosis by blocking transcription on damaged DNA templates. Mol Cancer Res: MCR 1, 747–754 (2003).
Yagi, T., Yarosh, D. B. & Day, R. S., 3rd. Comparison of repair of O6-methylguanine produced by N-methyl-N’-nitro-N-nitrosoguanidine in mouse and human cells. Carcinogenesis 5, 593–600 (1984).
Nonaka, M., Tsuchimoto, D., Sakumi, K. & Nakabeppu, Y. Mouse RS21-C6 is a mammalian 2′-deoxycytidine 5′-triphosphate pyrophosphohydrolase that prefers 5-iodocytosine. FEBS J 276, 1654–1666, doi: 10.1111/j.1742-4658.2009.06898.x (2009).
Konca, K. et al. A cross-platform public domain PC image-analysis program for the comet assay. Mutat Res 534, 15–20, doi: 10.1016/s1383-5718(02)00251-6 (2003).
Acknowledgements
The authors thank Masato Tanaka, Kaori Nagatoshi and Yuki Matsuzaki from the Laboratory for Technical Support, Medical Institute of Bioregulation, Kyushu University for their technical support regarding mouse embryo manipulation and DNA sequencing, Naomi Adachi, Kaoru Nakabeppu, Kazumi Asakawa and Tsukasa Kuwano for their technical assistance, Professor Mutsuo Sekiguchi and Dr. Riyoko Ito from Fukuoka Dental College for backcrosssing Mlh1 KO mice for ten generations, Dr. Hisaya Kawate from Nakamura Gakuen University for the preparation of anti-mouse MSH2 antibody, and Dr. Feng Zhang from the Massachusetts Institute of Technology for plasmid pX459 for the CRISPR/Cas9 system. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan [23117713 to DT] and from the Japan Society for the Promotion of Science [22221004 to YN].
Author information
Authors and Affiliations
Contributions
Y.Y. and D.T. wrote the manuscript, designed and performed major experiments; N.A. and T.I. contributed Figures 1, 2, 3, 4 and 5; N.S., M.M. and T.S. contributed the establishment of H414 cell line; K.S. and T.N. contributed Figure 5, Tables S1 and S4; Y.N. wrote the manuscript and designed experiments. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Yoneshima, Y., Abolhassani, N., Iyama, T. et al. Deoxyinosine triphosphate induces MLH1/PMS2- and p53-dependent cell growth arrest and DNA instability in mammalian cells. Sci Rep 6, 32849 (2016). https://doi.org/10.1038/srep32849
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep32849
- Springer Nature Limited
This article is cited by
-
An ITPA Enzyme with Improved Substrate Selectivity
The Protein Journal (2024)