Abstract
No coding sequence variants of the ALOX5AP gene that lead to amino acid substitutions have been identified. A two-stage study design was used to explore the relationship between variants in the transcriptional regulatory region of ALOX5AP gene and ischemic stroke (IS) risk in Chinese populations. IS was determined using CT and/or MRI. First, 18 SNPs, located in the upstream promoter region of ALOX5AP gene, were genotyped in 200 IS patients and 200 controls. And one potential associated SNP (rs17222919) was identified (P = 0.005,OR = 0.623, 95% CI: 0.448~0.866). Next, another independent case-control cohort comprising 810 IS patients and 825 matched controls was recruited to investigate the role of rs17222919, rs9579646 polymorphisms and their haplotypes in IS risk. The G allele frequency of rs17222919 in the IS group was significantly lower than that in control group (P = 0.007, OR = 0.792, 95% CI: 0.669~0.937). T-A and G-A haplotypes were associated with IS (P = 0.001,OR = 1.282, 95% CI:1.100~1.495; P = 0.0001, OR = 0.712, 95% CI: 0.598~0.848; respectively). Our study providesevidence that rs17222919 is a potential genetic protective factor against IS. Furthermore, the T-A haplotype is a risk factor and the G-A haplotype is a protective factor against IS in Chinese population.
Similar content being viewed by others
Introduction
Ischemic stroke (IS), also called cerebral infarction (CI), is a complex multifactorial disorder characterized by the sudden loss of blood circulation to an area of the brain, resulting in a corresponding loss of neurologic function1. Previous studies have suggested that inflammation is a key element in all critical steps of atherosclerosis, which underlies the pathogenesis of cardiovascular disease and IS2. The 5-lipoxygenase-activating protein (FLAP), which is encoded by the ALOX5AP gene, is a crucial regulator of the biosynthesis of leukotrienes (LTs), which can lead to the accumulation of LTs in fatty deposits on the arterial wall3. LTs initiate leukocyte activation and promote the adhesion of monocytes on the vascular wall, a process that plays an important role in the pathogenesis of atherosclerosis and inflammatory diseases, including IS4.
To date, most genetic studies have focused on the relationship between two at-risk haplotypes (HapA and HapB) of the ALOX5AP gene and the susceptibility to IS. However, the association results were inconsistent and controversial across different ethnic backgrounds5,6,7. In particular, no coding sequence variants of the ALOX5AP gene that lead to amino acid substitutions have been identified8. Domingues-Montanari et al.9 reported that the SG13S114 genotypes modulate the mRNA levels of ALOX5AP gene and the mRNA levels were higher in IS cases than in controls. Helgadottir et al.10 revealed that the ALOX5AP gene expression levels and its downstream leukotriene B4 (LTB4) synthesis activity were greater in IS patients than in controls. Kim et al.11 previously reported that a promoter polymorphism (rs17222919) was associated with the development of intracerebral hemorrhage in the Korean population. Ji RJ et al.12, also reported that another promoter polymorphism (−581_582 Ins A) might be a novel genetic risk factor for IS in a north Chinese Han population. These phenomena strongly suggested that there might be some additional unidentified variants in the regulatory regions of the ALOX5AP gene may play modulate IS risk. However, the relationship between polymorphisms throughout the entire transcriptional regulatory region of the ALOX5AP gene and IS risk has not been extensively explored.
Therefore, a two-stage study design was used to explore the relationship between variants of ALOX5AP gene and IS risk in two independent Chinese Han cohorts. Firstly, we selected 18 SNPs that cover the promoter region of the AlOX5AP gene to screen the positive SNPs using SNaPshot minisequence technique. Subsequently, we investigated the role of rs17222919 (in the promoter region) and rs9579646 (in the first intron region) polymorphisms in IS risk using TaqMan-PCR technique in a larger cohort from the Chinese Han population.
Results
Subject characteristics
The clinical and demographic characteristics of the two populations are shown in Table 1. Cases and controls were well matched in age and sex (P > 0.004; P > 0.025). Compared with the control groups, the IS groups showed higher percentages of hypertension, diabetes mellitus and smoking (P < 0.004; P < 0.025). IS patients also had significantly higher total cholesterol (TC) and total triglycerides (TG) levels than the control subjects ((P < 0.004; P < 0.025).
The preliminary screening results in the initial study
Association analysis of 18 SNPs in the promoter region of ALOX5AP with IS
The −1785G>A, −946A>G, −581_582lnsA, −519G>A, −290G>A and −190G>A were not polymorphic in our initial study. The genotype distributions of rs12560847, rs9578195, rs61947373, rs34404999, rs55950839, rs55780307, rs59227506, rs34536374, rs34344566, rs9578194 and rs34352240 were consistent with the Hardy–Weinberg equilibrium, but showed no significant differences between IS patients and controls (P > 0.004).
For the rs17222919 polymorphism, the G allele frequency was significantly lower in IS group (19.5%) than in control group (28.0%) (P = 0.005; OR = 0.623, 95% CI: 0.448~0.866). See Table 2.
Haplotype analysis of the ALOX5AP promoter region
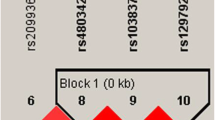
After the non-polymorphic loci were removed, linkage disequilibrium (LD) test was performed and the results were shown in Fig. 1. The rs34536374, rs34344566, rs34352240, rs9578194, rs55950839, rs55780307, rs59227506 and rs12560847 were in strong linkage disequilibrium and haplotype blocks (D’ > 0.8) were defined. Because rs17222919 was not in strong linkage disequilibrium (D’<0.8), the haplotype blocks were defined without rs17222919. The distribution of haplotype (CACGTATG) and haplotype (TGTAAGCA) has no significant difference between IS group and control group (P = 0.037). See Table 3.
Association between the rs17222919 and rs9579646 polymorphisms and IS in the second cohort
Association analysis and inherited model test
The genotype frequency distributions of rs17222919 and rs9579646 were consistent with Hardy-Weinberg equilibrium (HWE) in the second cohort. After adjusting for conventional risk factors, the G allele frequency (19.3%) in the IS group was found to be significantly lower than that (23.2%) in the control group (P = 0.007, OR = 0.792, 95% CI = 0.669~0.937). See Table 4. However, the rs9579646 polymorphism showed no significant differences between the two groups (P > 0.025).To assess the effect of rs17222919 on the risk of IS, we compared additive, dominant, and recessive models. The effect of rs17222919 was best described with the dominant model (P = 0.016 OR = 0.778, 95% CI = 0.637~0.951). See Table 5.
Linkage disequilibrium test and haplotype analysis
Linkage disequilibrium test indicated that rs17222919 and rs9579646 were in strong linkage disequilibrium (D’ = 0.837). Consequently, haplotype analysis was carried out. Three common haplotypes (frequency >3%) in the cases and controls were presented in Table 6. There was a statistically significant difference in the frequency of the G-A haplotype between the case and control groups (P = 0.0001, OR = 0.712, 95% CI: 0.598–0.848). The frequency of the T-A haplotype was higher in the IS group than in the control group (P = 0.001, OR = 1.282, 95% CI: 1.100~1.495).
Discussion
Promoter elements have fundamental roles in regulating gene transcription. Altering the DNA sequence of a promoter may result in changes in transcription factor binding sites or binding rates. Thus, a single nucleotide polymorphism (SNP) in the promoter region can affect promoter activity by altering the binding affinity of transcription factors involved in the regulation of gene expression13,14. In the present study, we designed a two-stage study to explore the relationship between variants in the transcriptional regulatory region of the ALOX5AP gene and IS risk. To the best of our knowledge, this is a comprehensive study to evaluate whether polymorphisms in the transcriptional regulatory region of ALOX5AP gene could influence susceptibility to IS in a large Han Chinese population.
Firstly, 18 SNPs that cover the promoter region of the ALOX5AP gene were selected using SNaPshot method to screen positive SNPs. The results showed that −1785G>A, −946A>G, −581_582lnsA, −519G>A, −290G>A and −190G>A loci were not polymorphic in our initial population. Our results were obviously different from those reported by Ji RJ et al.12, who reported that three genetic variants were identified, including a mutation (−519 G/A), an insertion and deletion polymorphism (−581_582 Ins A) and a single nucleotide polymorphism (−946 A/G). They also reported that the −581_582 Ins A polymorphism might be a novel genetic risk factor for IS in a north Chinese Han population. The discrepancy between their findings and those of our study may be attributed to the differences in sample size. Therefore, further study with a larger population is required to confirm the findings. Next, the genotype distributions of 11 SNPs, including rs12560847, rs9578195, rs61947373, rs34404999, rs55950839, rs55780307, rs59227506, rs34536374, rs34344566, rs9578194 and rs34352240 were consistent with the Hardy–Weinberg equilibrium but showed no significant differences between IS and controls (P > 0.004). However, the G allele frequency of rs17222919 was significantly different between the IS and control groups (P = 0.005; OR = 0.623, 95% CI: 0.448~0.866). If the adjusted P value for significance was set at 0.05/12 = 0.004 in the initial cohort by Bonferroni’s adjustment, the P value of rs17222919 (0.005) was close to the statistical significance. So we thought it was a potentially associate locus.Therefore, the rs17222919 was identified to have a potentially positive association with IS in the first-stage study and was to be replicated in a larger Chinese Han cohort.
Haplotype analysis is considered more powerful than single SNP analysis to search for genetic determinants of complex diseases15. After the non-polymorphic loci were excluded, LD test was performed. The results indicated that rs34536374, rs34344566, rs34352240, rs9578194, rs55950839, rs55780307, rs59227506 and rs12560847 were in strong linkage disequilibrium (D’ > 0.8) and haplotype blocks were defined. The 8 SNPs constituted a total of 22 common potential haplotypes in the patients and controls. And the most frequent haplotype was Hap(CACGTATG). The frequency of haplotype (CACGTATG) was significantly higher in the IS patients than in controls (P = 0.037,OR = 1.540, 95% CI:1.024~2.314). The frequency of haplotype (TGTAAGCA) was significantly lower in the IS patients than in controls (P = 0.037,OR = 0.650, 95% CI:0.432~0.976). However, both of the two haplotypes didn’t reach the statistical significance. Further study with a larger population is required to evaluate the findings.
Preliminary studies have revealed that the introns have important biological functions. In particular, the first introns of human genes are likely to be involved in transcriptional regulation16,17. Thus, in the second larger Chinese cohort, we investigated the role of rs17222919 (in the promoter region) and rs9579646 (in the first intron region) polymorphisms in IS risk using TaqMan-PCR technique. For the rs17222919 polymorphism, the G allele frequencies of IS group (19.3%) were significantly lower than in the control group (23.2%) (P = 0.007). Multivariate logistic regression analysis showed that the G allele was associated with a 0.792-fold increased risk of IS after adjusting for conventional risk factors (95% CI, 0.669~0.937; P = 0.007). Rs17222919 was associated with IS in a dominant genetic model (P < 0.025) after performing genotype association tests with dominant, recessive and additive models. However, this association was inconsistent with the results previously reported by Kim et al.11, who showed that rs17222919 was associated with intracerebral hemorrhage, but not IS, in a Korean population. Our previous in vitro promoter assay revealed that the G allele had a lower transcriptional activity than the T allele, suggesting that the −1316T/G variation reduces ALOX5AP promoter activity and consequently down-regulates gene expression. The decreased ALOX5AP transcription results in increased inactivation of the 5-LO pathway and reduces leukotriene biosynthesis, which in turn protects from IS18. The rs9579646 polymorphism showed no significant differences between groups and was not associated with IS in our second Chinese cohort.
Finally, the linkage disequilibrium test and haplotype analysis were performed using SHEsis software. The rs17222919 and rs9579646 polymorphisms were in complete linkage disequilibrium (D’ = 0.837). The frequency of the G-A haplotype was lower in the IS group than in the controls (P = 0.0001), suggesting that the G-A haplotype might be a genetic protective factor against IS in the Chinese Han population (OR = 0.712, 95% CI: 0.598–0.848). Conversely, the T-A haplotype was associated with an increased risk of IS (OR = 1.282, 95% CI:1.100~1.495) and the T-A haplotype might be a risk factor for IS in this Chinese Han cohort. Furthermore, the G-G haplotype frequencies were quite low, and this haplotype thus had less impact on the incidence of IS.
Several limitations of our study need to be addressed. First, the sample size of the initial study (200 cases and 200 controls) may not be sufficiently large to screen all the potential risk-associated SNPs and evaluate gene–environment interactions. Second, there was potential selection bias because the cases and controls were recruited from hospital. Third, ALOX5AP mRNA levels among different rs17222919 genotypes were not compared in either the IS or control group.
In conclusion, we designed a two-stage study to explore the relationship between variants in transcriptional regulatory region of the ALOX5AP gene and IS. In the first stage, 18 SNPs covering the promoter region of the ALOX5AP gene were screened and one potential risk-associated SNP (rs17222919) was selected. In the second larger Chinese cohort, we confirmed that the rs17222919 polymorphism was associated with a decreased risk of IS. In addition, two haplotypes were discovered to be associated with IS. Future studies of the variants in the transcriptional regulatory region of ALOX5AP and their biological functions should be conducted to further elucidate the etiology of IS.
Methods
Study populations
In the initial study, 200 IS patients (112 men and 88 women, mean age 57.2 ± 7.2 years) were recruited from the First Affiliated Hospital of Zhengzhou University. In the second cohort, a total of 810 patients with ischemic stroke (males: females = 416:394, mean age 57.7 ± 8.6 years) were enrolled from Henan Provincial Hospital in central China. None of the participants were included in both populations. The IS was defined by a loss of global or focal cerebral function persisting for >24 h with corresponding infarction on brain imaging with a probable vascular cause19. IS cases were classified into three subtypes, namely large-artery atherosclerosis (LAA), small-artery occlusion lacunar (SAO), and stroke of other undetermined etiology (SUE), according to the Trial of Org 10172 in Acute Stroke Treatment (TOAST)20. Brain imaging was carried out using computed tomography (CT) and/or magnetic resonance imaging (MRI) as well as ancillary diagnostic investigations and standardized blood tests were also performed. Patients with atrial fibrillation, cerebral hemorrhage, peripheral vascular diseases, or kidney diseases were excluded from the study.
The control groups consisted of 200 (initial study: 106 men and 94 women, mean age 55.9 ± 7.0 years) and 825 (second study: 428 men and 397 women, mean age 55.3 ± 7.2 years) unrelated Henan Han individuals, selected from the same demographic area and matched to the cases by age, sex, and residency. All controls were free of cerebrovascular disease, cardiovascular disease, hepatic disease, renal disease and cancer.
The study protocols were approved by the Ethics Committee on Human Research of Zhengzhou University and informed written consent was obtained from each participant. All experiments were performed in accordance with relevant guidelines and regulations.
Preliminary screening for positive SNPs by SNaPshot
Because promoter elements have key roles in regulating gene transcription, we selected 18 SNPs covering the promoter region of ALOX5AP gene for initial analysis. The −1785G>A, −946A>G, −581_582lnsA, −519G>A, −290G>A and −190G>A were selected based on previously reported significant associations. The rs12560847, rs9578195, rs61947373, rs17222919, rs34404999, rs55950839, rs55780307, rs59227506, rs34536374, rs34344566, rs9578194 and rs34352240 were selected based on pairwise r2 (>0.8) among all common SNPs with minor allele frequency (MAF) >0.1 spanning the promoter region of the AlOX5AP gene using the Haploview 4.0 software21.
Genomic DNA was extracted from peripheral white blood cells of 200 pairs case and control participants using the Blood Genomic DNA Miniprep Kit (Axygen Biotechnology, Union City, CA, USA) according to the manufacturer’s instructions. Touch-down PCR amplifications was performed using 7 pairs of primers (Sangon, Shanghai, China). All primers were designed using the primer3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). All samples were genotyped using SNaPshot minisequence technique.
A 10 μL mixture containing 1x HotStarTaq buffer, 3.0 mM Mg2+, 0.3 mM dNTP, 1U HotStarTaq polymerase (Qiagen), 1 μL multiple PCR primers and 1 μL template DNA was prepared for each reaction. The cycling program was 95 °C for 2 mins; 11 cycles of 94 °C for 20 s, (62 °C −0.5 °C/cycle) for 40 s, 72 °C for 1.5 mins; 24 cycles of 94 °C for 20 s, 57 °C for 30 s, 72 °C for 1.5 mins; and 72 °C for 2 mins. To eliminate excess primers and dNTPs, 5U SAP (Promega, USA) and 2U Exo I (Epicentre, Palmerston North, New Zealand) were added to the 10 μL PCR products. The mixture was incubated at 37 °C for 60 mins, followed by incubation at 75 °C for 15 mins.
Eighteen extension primers were designed for multiplex PCR SNaPshot reaction. The reaction mixture included 5 μL SNaPshot Multiplex reaction mix (Applied biosystems, USA), 2 μL ddH2O, 1 μL extension primer mix and 2 μL purified PCR product. The cycling program was 96 °C for 1 min; 28 cycles of 96 °C for 10 s, 55 °C for 5 s and 60 °C for 30 s. In order to purify the extension products, 1U SAP (Promega, USA) was added to extension product and incubated at 37 °C for 60 mins, followed by incubation at 75 °C for 15 mins. Finally, a mixture of 9 μL HiDi Formamide, 0.5 μL Liz120 size standard and 1 μL purified extension product were denatured at 95 °C for 5 mins and then loaded onto an ABI3730xl instrument (ABI, USA) and GeneMapper4.1 (Applied biosystems, USA) was run to analyze the results.
Genotyping of rs17222919 and rs9579646 polymorphisms
The preliminary screening results showed that only rs17222919 in the ALOX5AP gene promoter region had a significantly different frequency in the IS and control groups. Moreover, the first introns of human genes are likely to be involved in transcriptional regulation. Thus, in the second cohort, we selected rs17222919 which was positively associated with IS in the preliminary study and rs9579646 which is located in the first intron of the ALOX5AP gene for analysis.
EDTA anti-coagulated venous blood samples were collected from the 810 enrolled IS patients and 825 healthy controls. Genomic DNA was extracted from the peripheral blood using the Blood Genomic DNA Miniprep Kit (Axygen Biotechnology, Union City, CA, USA). TaqMan probes were used to analyze rs17222919 and TaqMan-MGB probes were selected to genotype rs9579646 polymorphism respectively. The genotype was determined according to the relative fluorescence intensity of the probe detected by the real-time PCR system (ABI PRISM 7500; Applied Biosystems, Foster City, CA, USA). Each PCR reaction mixture (10 μL) contained 5 μL of 2× TaqMan Universal PCR Master Mix (Applied Biosystems), 0.2 μL forward primer, 0.2 μL reverse primer, 0.6 μL FAM-labeled probe, 0.6 μL HEX-labeled probe, 0.4 μL ROXII, 1.0 μL of DNA template (1~20 ng/μL), and 2 μL ddH2O. In each assay, three samples with known genotypes and three no-DNA blank controls were included. The PCR reaction conditions consisted of pre-degeneration for 2 mins at 95 °C, followed denaturation at 95 °C for 15 s, and 40 cycles of annealing and extension at 60 °C for 30 s. After PCR amplification, sample genotypes were determined by measuring the allele-specific fluorescence with an ABI Prism 7700 Sequence Detection System and using SDS 1.7 software for allele discrimination (Applied Biosystems). To verify the genotyping accuracy, 10% of random samples were sequenced.
Statistical analysis
All statistical analysis was performed using the SPSS 17.0 package (SPSS Inc., Chicago, IL, USA). Pearson’s chi-squared test was used to test for differences in qualitative variables and genotype/allele frequencies. Differences in quantitative variables between groups were analyzed using Student’s t-test. Testing for deviation of genotype distribution from Hardy–Weinberg equilibrium and a haplotype-based case-control study were performed using SHEsis software (http://analysis.bio-x.cn)22. Odds ratios (ORs), 95% confidence intervals (95% CIs) and corresponding P values for IS risk were calculated by logistic regression analysis after adjusting for age, gender, hypertension, diabetes, smoking, drinking and biochemical indexes such as TC and TG levels. Bonferroni’s adjustment was used for multiple comparisons. The adjusted P value for significance was set at 0.05/12 = 0.004 in the initial cohort and the adjusted P value less than 0.025 were considered statistically significant in the second cohort.
Additional Information
How to cite this article: Yang, D. et al. Genetic Variants in the Transcriptional Regulatory Region of the ALOX5AP gene and Susceptibility to Ischemic Stroke in Chinese Populations. Sci. Rep. 6, 29513; doi: 10.1038/srep29513 (2016).
References
Mirzaei, M. et al. Cerebrovascular disease in 48 countries: secular trends in mortality 1950–2005. J Neuro Neurosur Ps 83, 138–145 (2012).
Zintzaras, E. et al. Variants of the Arachidonate 5-Lipoxygenase-Activating Protein (ALOX5AP) Gene and Risk of Stroke: A HuGE Gene-Disease Association Review and Meta-Analysis. Am J Epidemiol 169, 523–532 (2012).
Evans, J. et al. What’s all the FLAP about?: 5-lipoxygenase-activating protein inhibitors for inflammatory diseases. Trends Pharmacol Sci 29, 72–78 (2008).
Spanbroek, R. et al. Expanding expression of the 5-lipoxygenase pathway within the arterial wall during human atherogenesis. Proc Natl Acad Sci USA 100, 1238–1243 (2003).
Bisoendial, R. J. et al. The association between the gene encoding 5- lipoxygenase activating protein and abdominal aortic aneurysms. Atherosclerosis 220, 425–428 (2012).
Papapostolou, A. et al. Are ALOX5AP gene SNPs a risk or protective factor for stroke? Gene 548, 56–60 (2014).
Yang, D. et al. A Novel Risk Haplotype of ALOX5AP Gene is Associated with Ischemic Stroke in Chinese Han Population. J Mol Neurosci 53, 493–499 (2014).
Yi, X. et al. Genetic Polymorphisms of ALOX5AP and CYP3A5 Increase Susceptibility to Ischemic Stroke and Are Associated with Atherothrombotic Events in Stroke Patients. J Stroke Cerebrovasc 24, 521–529 (2015).
Domingues-Montanari, S. et al. Association of a Genetic Variant in the ALOX5AP with Higher Risk of Ischemic Stroke: A Case-Control, Meta- Analysis and Functional Study. Cerebrovasc Dis 29, 528–537 (2010).
Helgadottir, A. et al. The gene encoding 5-lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nat Genet 36, 233–239 (2004).
Kim, D. H. et al. A Promoter polymorphism (rs17222919, –1316T/G) of ALOX5AP is associated with intracerebral hemorrhage in Korean population. Prostag Leukotr Ess 85, 115–120 (2011).
Ji, R. et al. Genetic Variants in the Promoter Region of the ALOX5AP Gene and Susceptibility of Ischemic Stroke. Cerebrovasc Dis 32, 261–268 (2011).
Yocum, G. T. et al. Inducible Nitric Oxide Synthase Promoter Polymorphism Affords Protection Against Cognitive Dysfunction After Carotid Endarterectomy. Stroke 40, 1597–1603 (2009).
Chen, C. et al. An NKX3.1 binding site polymorphism in the l-plastin promoter leads to differential gene expression in human prostate cancer. Int J Cancer, n/a-n/a (2015).
Ni. Y. et al. The single nucleotide polymorphism and haplotype analysis of MDR1 in Chinese diffuse large B cell lymphoma patients. Biomed Pharmacother 73, 24–28 (2015).
Li, H. et al. Analysis of intron sequence features associated with transcriptional regulation in human genes. PLoS One 7, e46784 (2012).
Ying, S. et al. An Intronic Enhancer Driven by NF-jB Contributes to Transcriptional Regulation of Peptidylarginine Deiminase Type I Gene in Human Keratinocytes. J Invest Dermatol 130, 2543–2552 (2010).
Fan, Y. et al. A Promoter Polymorphism (rs17222919, −1316T/G) of ALOX5AP Gene Is Associated with Decreased Risk of Ischemic Stroke in Two Independent Chinese Populations. PLoS One 10, e122393 (2015).
Saleheen, D. et al. Association of Phosphodiesterase 4D Gene With Ischemic Stroke in a Pakistani Population. Stroke 36, 2275–2277 (2005).
Adams, H. P. et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke 24, 35–41 (1993).
Barrett, J. C. et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Shi, Y. et al. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 15, 97–98 (2005).
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 81571154) and by grants from the Leading Talent Project of the Zhengzhou City Science and Technology Bureau (project number 131PLJRC679). We acknowledge the technical assistance of staff members of the Neurology Department of the First Affiliated Hospital of Zhengzhou University. We also thank all patients and controls for providing blood samples.
Author information
Authors and Affiliations
Contributions
Y.Z. and Y.L. performed experiments. X.H. conducted the statistical analysis. C.C. and X.Z. were involved in local study implementation and participant recruitment. D.Y. and Y.H. wrote the manuscript. Y.H. and H.Z. conceived of the study, and participated in its design and coordination. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Yang, D., Huang, X., Cui, C. et al. Genetic Variants in the Transcriptional Regulatory Region of the ALOX5AP gene and Susceptibility to Ischemic Stroke in Chinese Populations. Sci Rep 6, 29513 (2016). https://doi.org/10.1038/srep29513
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep29513
- Springer Nature Limited