Abstract
Acaryochloris marina, a unicellular oxygenic photosynthetic cyanobacterium, has uniquely adapted to far-red light-enriched environments using red-shifted chlorophyll d. To understand red-light use in Acaryochloris, the genome of this cyanobacterium was searched for red/far-red light photoreceptors from the phytochrome family, resulting in identification of a putative bacteriophytochrome AM1_5894. AM1_5894 contains three standard domains of photosensory components as well as a putative C-terminal signal transduction component consisting of a histidine kinase and receiver domain. The photosensory domains of AM1_5894 autocatalytically assemble with biliverdin in a covalent fashion. This assembled AM1_5894 shows the typical photoreversible conversion of bacterial phytochromes with a ground-state red-light absorbing (Pr) form with λBV max[Pr] 705 nm, and a red-light inducible far-red light absorbing (Pfr) form with λBV max[Pfr] 758 nm. Surprisingly, AM1_5894 also autocatalytically assembles with phycocyanobilin, involving photoreversible conversion of λPCB max[Pr] 682 nm and λPCB max[Pfr] 734 nm, respectively. Our results suggest phycocyanobilin is also covalently bound to AM1_5894, while mutation of a cysteine residue (Cys11Ser) abolishes this covalent binding. The physiological function of AM1_5894 in cyanobacteria containing red-shifted chlorophylls is discussed.
Similar content being viewed by others
Introduction
Photoreceptors detect changes in light quality, and in response, they trigger cellular signals that regulate downstream physiological processes. Phytochromes are a group of photoreceptors that are particularly important to photosynthetic organisms because they detect photosynthetically active radiation, having absorption maxima similar to chlorophylls1. Phytochromes, which were first characterized in plants, act as red/far-red photoreceptors in two interconvertible forms: a red light-absorbing form (Pr) and a far-red light-absorbing form (Pfr). The ground state Pr form converts to the activated state Pfr upon illumination with red light. Extended dark or illumination with far-red light converts the Pfr form back to the Pr form. Phytochromes have since been identified in fungi, photosynthetic and non-photosynthetic bacteria2.
Phytochromes are modular proteins that typically consist of an N-terminal photosensory core composed of PAS (a domain named after the proteins: period circadian protein, aryl hydrocarbon receptor nuclear translocator protein, and single-minded protein); GAF (a domain, named after cGMP-specific phosphodiesterases, adenylyl cyclases, and FhlA proteins) and PHY (phytochrome) domains, as well as a C-terminal signal transduction domain that is commonly a histidine kinase3. The phytochrome superfamily has been subdivided into a number of clades: plant phytochromes (Phys), cyanobacterial phytochromes (Cphs), bacteriophytochromes (BphPs), fungal phytochromes (Fphs) and the more distantly related Phy-like phytochromes (also known as cyanobacteriochromes or CBCRs)2,4. The binding of different linear tetrapyrroles (or bilins), as well as their manner of binding, allows phytochromes from different organisms to detect changes in light conditions relevant to a particular organism. All bilins are derived from a heme molecule, converted to the linear tetrapyrrole biliverdin IXα (BV) by heme oxygenase. This BV can be modified to 3-E-phytochromobilin (PΦB) or 3-Z-phycocyanobilin (PCB) by bilin reductases. While BphPs typically bind the simplest linear tetrapyrrole (BV) via a conserved cysteine (Cys) residue in the PAS domain in vivo5, Phys and Cphs bind PΦB and PCB, respectively, via a Cys residue in the GAF domain6. In addition to Cphs, some cyanobacteria possess cyanobacterial bacteriophytochromes (cBphPs). These cBphPs are phylogenetically similar to Cphs, but like BphPs they have a conserved Cys residue in their PAS domain, and lack a chromophore-binding Cys residue in their GAF domain2. There is evidence to support that a highly conserved His residue in the GAF domain is important for chromophore attachment in some BphPs7. However, crystallographic data suggest that this highly conserved His residue is involved in a network of hydrogen bonding in the chromophore-binding pocket8, rather than acting as a covalent ligation site. Atypically, some BphPs have their ground state absorption maxima at a longer wavelength than their light-activated state (i.e., the ground state is the Pfr form), and are called bathyphytochromes (Bathy BphPs) accordingly5,9,10.
CBCRs are the most structurally diverse subgroup of the phytochromes, often missing highly conserved amino acid residues found in other phytochromes; they can respond to nearly the entire visible light spectrum11. CBCRs often contain multiple GAF domains with Cys residues, which can either reversibly or irreversibly bind PCB1,4,12,13. The evolution of such a diverse family of photoreceptors in cyanobacteria as well as the wide spectral range detected by these photoreceptors highlights the importance of light quality for cyanobacteria. Clearly, this reflects the fact that cyanobacteria are photoautotrophs that harvest light for photosynthesis at a number of different wavelengths, the amounts of which are dependent on the level they inhabit in a water column and competition from other photosynthetic organisms14. Recent research has demonstrated that phytochromes from eukaryotic algae that inhabit similar ecological niches to cyanobacteria also have evolved complicated photocycles, beyond typical red/far-red light switching mechanisms15.
Acaryochloris marina (Acaryochloris) is a cyanobacterium that uses chlorophyll d (Chl d) instead of Chl a in the reaction centre of both photosystems16,17. Recently, two additional cyanobacterial species have been shown to contain trace amounts of Chl d18,19. However, the function of Chl d in these organisms has not been established. The type species, Acaryochloris MBIC11017, was discovered as a symbiont of a colonial ascidian in the Pacific Ocean20. Subsequently, several ecotypes of Acaryochloris have been discovered in niche environments around the world that are enriched in red/far-red light21,22,23,24,25,26,27,28. Chl d has a red-shifted absorbance29, which allows Acaryochloris to exploit such niche environments22. The QY peaks of Chl a and Chl d in vivo are approximately 675 nm and 712 nm, respectively30, giving Chl d an absorption maximum intermediate to Chl a and bacteriochlorophylls31. Acaryochloris captures light energy in its reaction centres using two discrete light-harvesting systems: (1) a Chl d-binding antenna predominantly of Chl d with small amounts of Chl a32; and (2) a phycobiliprotein antenna that is a simpler version of phycobilisomes found in other cyanobacteria33,34. Acaryochloris is able to regulate these antennas in response to changing light quality or intensity in a manner similar to complementary chromatic adaptation30,35,36. The newly identified far-red/green reversible cyanobacteriochrome AM1_1870 indicates that Acaryochloris has a complementary light regulatory system to different environmental conditions37. In addition, a number of characterised CBCRs with photocycles ranging from blue to far-red absorption maxima (up to absorption peak at 706 nm) suggest Acaryochloris may use these to signal responses to different light conditions38,39.
In this work, a putative phytochrome gene, AM1_5894, was identified in the Acaryochloris genome using BLAST and sequence analysis40. Sequence comparison revealed that AM1_5894 is part of a small cluster of cyanobacterial BphPs, which are related to BphPs from non-photosynthetic and anoxygenic photosynthetic bacteria, but distinct from the typical Cphs in Chl a-containing cyanobacteria. We demonstrate that heterologously expressed AM1_5894 covalently binds the chromophores PCB and BV, and undergoes photoconversion between a Pr and Pfr form in vitro. The reconstituted BV-AM1_5894 has a spectral profile in its Pr/Pfr state (λBV max[Pr] 705 nm and λBV max[Pfr] 758 nm). Reconstitution of AM1_5894 with PCB blue shifts the absorption maxima by approximately 23 nm (λPCB max[Pr] 682 nm and λPCB max[Pfr] 734 nm), broadening the AM1_5894 light detection range, when it acts as a photoreceptor in Acaryochloris. Furthermore, covalent attachment of both chromophores is abolished when the presumed adduct (Cys11) is mutated. We show that it is advantageous for Acaryochloris to have a BphP instead of a typical Cph protein in its enriched red/far-red light-enriched environment.
Results
Protein domain comparison
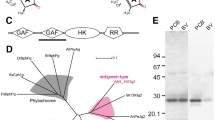
The AM1_5894 gene comprises 2544 nucleotides and encodes an 847-amino acid protein product, having a predicted molecular weight of 94.2 kDa. The overall domain structure of AM1_5894 is typical of other bacteriophytochromes, with a photosensory core module of PAS-GAF-PHY as well as a C-terminal signal transduction domain, consisting of a histidine kinase; and a receiver domain (Fig. 1). The PAS domain contains a Cys residue at its eleventh position that aligns with the conserved BV-binding Cys of typical BphPs. It also has a His residue (H249) in its GAF domain that is conserved (Fig. 1). The adjacent PCB/PΦB-binding Cys found in the GAF domain of cyanobacterial and plant phytochromes is absent in AM1_5894 (Fig. 1). AM1_5894 is more closely related to BphPs from non-photosynthetic and anoxygenic photosynthetic bacteria than cyanobacterial phytochromes (Cph), based on alignment of their GAF domains40. Extending this alignment to include both the GAF and PHY domains confirms that AM1_5894 is phylogenetically separate from other Cphs and cBphPs, apart from the BphPs from another Acaryochloris ecotype, CCMEE 5410, and the filamentous cyanobacterium Leptolyngbya PCC 7375 (Figure S1). Unusually, the signal output domain of this cluster of three BphPs consists of a HWE (histidine-tyrosine-glutamate)-type histidine kinase and a receiver domain, which is a typical domain structure of Bathy BphPs. Bathy BphPs are most commonly found in Rhizobiales10,41. Although Acaryochloris MBIC 11017 does not contain any Cph-like phytochromes, a number of putative CBCRs, including the recently characterized AM1_1186, AM1_1870 and AM1_1557, are present37,38,39.
The domain structure of AM1_5894 and the truncated AM1_5894ΔHK used in this study are shown. Alignments highlight the conserved Cys residue in the PAS domain (open triangle; named after period circadian protein, aryl hydrocarbon receptor nuclear translocator protein and single-minded protein) that covalently binds BV in bacteriophytochrome (BphP) and cyanobacterial BV-binding bacteriophytochrome (cBphP) and the conserved Cys residue in the GAF domain (closed triangle; named after cGMP-specific phosphodiesterases, adenylyl cyclases and FhlA proteins) that binds PCB or phytochromobilin (PΦB) in proteins in cyanobacterial phytochromes (Cphs) and plant phytochromes (Phys), respectively. The his residue adjacent this position which is conserved in most phytochromes is also marked (closed circle). HWE (histidine-tyrosine-glutamate -type histidine kinase(HK); HA_c (Histidine kinase-like ATPase c-terminal region); REC (cheY-like receiver domain). For other abbreviations refer to the text. Additional sequence information is given in Supplementary Table 1.
Expression of recombinant AM1_5894
Initial attempts to clone and express the full-length AM1_5894 were largely unsuccessful. A clone with a point mutation in its histidine kinase domain causing a Gly639 → Asp substitution was obtained, although only a small amount of soluble protein showing typical bacteriophytochrome spectral characteristics could be expressed in Escherichia coli (Figure S2). Instead, a truncated construct was expressed in E. coli. This truncated protein (AM1_5894∆HK) is 508-amino acids in length, compared with the full-length protein of 847 amino acids. AM1_5894∆HK contains PAS, GAF and PHY domains that are essential for chromophore attachment and photochromism42,43,44,45, but lacks the output module, including the histidine kinase and receiver domain (Fig. 1). AM1_5894∆HK was His-tag purified almost to homogeneity and used to examine the chromospectral properties of the AM1_5894 holoprotein.
Chromophore-protein Reconstitution
Based on sequence analysis, AM1_5894 was predicted to covalently bind BV, but not PCB because of a conserved Cys residue in its PAS domain, and lack of a conserved PCB/PΦB-binding Cys residue in its GAF domain (Fig. 1). To confirm this prediction, overexpressed full-length AM1_5894D639G and truncated AM1_5894∆HK were incubated in the absence or presence of either BV or PCB to facilitate autocatalytic attachment of the chromophore. Surprisingly, both reconstituted BV- and PCB-AM1_5894 protein complexes exhibited Zn-induced fluorescence on SDS-polyacrylamide gels (Fig. 2a). This demonstrates that both BV and PCB are covalently bound to AM1_5894 in vitro. Due to the low yields and point mutation of our full-length AM1_5894 clone, the truncated construct (AM1_5894∆HK) was used for chromospectral analyses of AM1_5894.
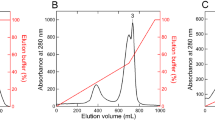
(a) SDS-polyacrylamide gel of either full length AM1_5894D639G or AM1_5894ΔHK after incubation in the absence (Apo) or presence of either BV or PCB. Covalently bound chromophore was detected by Zn induced fluorescence (Zn). Closed arrows mark the fluorescent chromophore-protein complexes. Asterisks mark degradation products of the full length AM1_5894D639G which were not observed for the truncated construct. CBB, Coomassie brilliant blue. (b) BioSec2000 purification of assembled AM1_5894ΔHK with BV and with PCB. The assembled chromophore protein complexes have a retention time of approximately 7.8 min (c) on-line spectral comparison of BV- and PCB-assembled AM1_5894ΔHK from the BioSec2000 column at indicated retention times.
In order to avoid any bias that may be generated by self-aggregated chromophores, either BV or PCB was added directly to lysed E. coli containing overexpressed AM1_5894∆HK before His-tag purification. Further purification was carried out using non-denaturing size exclusion chromatography (Fig. 2b). Online spectra of the chromophore-binding protein complexes confirmed the elution peak at 7.8 min corresponded to the assembled holoproteins, with the same spectra observed in vitro (Figs 2c and 3).
(a) Absorbance spectrum of the BV-AM1_5894ΔHK in its Pr state (solid line) and its Pr + Pfr mixed state (dashed line) following exposure to saturating 777 nm light. The difference spectrum (dash dotted line) is shown below. (b) Absorbance spectrum of the PCB-AM1_5894PΔHK in its Pr state (solid line) and its Pr + Pfr mixed state (dashed line) following exposure to saturating 777 nm light. The difference spectrum (dash dotted line) is shown below. Wavelength maxima and minima are indicated.
Spectral characterization
The absorption spectrum of dark-assembled BV-AM1_5894ΔHK resembles that of a typical phytochrome in its Pr state (Fig. 3a). It has two main absorption peaks centred at 380 and 705 nm, with a shoulder at approximately 650 nm (Fig. 3a). Upon illumination with red light (665 nm), there is a reduction in the 705 nm peak, accompanied by the appearance of a longer wavelength peak at 758 nm, which is indicative of the Pfr form (dashed line, Fig. 3a). Illumination of the Pfr form with far-red light (777 nm) or dark incubation causes AM1_5894 to switch back to its Pr state. The difference spectrum of BV-AM1_5894ΔHK Pfr minus Pr demonstrates a 378 nm minimum and 422 nm maximum peaks in the blue region, and 705 nm minimum and 758 nm maximum peaks in the far-red region (Fig. 3a). We found it was not possible to completely switch BV-AM1_5894ΔHK to its Pfr state, and the ratio between Pr and Pfr states remained stable after 15 min (Fig. 4a). The Pfr-state protein rapidly reverted back to its Pr state when exposed to far-red light (777 nm) (Fig. 4b) or incubated in the dark.
After dark assembly with the chromophore PCB, the AM1_5894ΔHK spectrum shares a similar, but blue-shifted peak structure to that of BV-AM1_5894ΔHK (Fig. 3). PCB-AM1_5894ΔHK has major peaks at 370 nm and 682 nm and a shoulder at approximately 625 nm (Fig. 3b). Illumination with red light initiates a partial switch to the Pfr state of PCB-AM1_5894ΔHK, with a maximum at 734 nm. The difference spectrum of Pfr minus Pr shows similar profiles and 368 nm minimum and 412 nm maximum peaks in the blue light region and 682 nm minimum and 734 nm maximum peaks in the red/far-red light region (Fig. 3b). By normalising at an absorbance of 280 nm, the absorbance intensity at 705 nm for BV-AM1_5894ΔHK is higher than that at 682 nm for PCB-AM1_5894ΔHK (Fig. 2c), indicative of a lower binding affinity of AM1_5894ΔHK for PCB compared with BV.
Like the BV-holoprotein, the PCB-holoprotein also could not be entirely switched to its Pfr state (Fig. 5). However, the switching kinetics between Pr-Pfr and Pfr-Pr were very different between BV-AM1_5894ΔHK and the PCB-AM1_5894ΔHK (Figs 4 and 5). The Pr-Pfr switching of BV-AM1_5894ΔHK was slow with a halflife of ~3.5 minutes while the Pfr-Pr switching was fast with a halflife of less than 5 sec (Fig. 4). Unlike the BV-AM1_5894ΔHK, the switching kinetics of PCB-AM1_5894ΔHK was approximately symmetric with one another, whether switching from Pr to Pfr or vice versa with a halflife of approximately 10 sec for the conversion (Fig. 5).
Differences in dark relaxation and photoactive light range
Dark relaxation and the effects of different light wavelengths on the switching of the Pfr form of AM1_5894 to its Pr state were examined in detail for BV-AM1_5894ΔHK and PCB-AM1_5894ΔHK using different wavelength light emitting diodes (LEDs) (Fig. 6a,b, Figure S3). Maximal Pfr to Pr switching for both BV- and PCB-bound AM1_5894ΔHK was observed when exposed to far-red light (777 nm), with a similar rate of switching observed for PCB-AM1_5894ΔHK exposed to 735 nm light. Dark relaxation was rapid (approximately 85% complete after 30 s) for BV-AM1_5894ΔHK, with rates similar to those observed when it was exposed to 777 nm light, while exposure to other light wavelengths inhibited this switching (Fig. 6a). In contrast, there was only minimal dark relaxation observed for PCB-AM1_5894ΔHK after 60 s. The dark reversion of BV-AM1_5894ΔHK is faster than PCB-AM1_5894ΔHK. The low ratio of bound PCB in reconstituted PCB-AM1_5894ΔHK complexes (Fig. 2c) and the slower dark reversion (Fig. 6) support the fact that, as predicted, the BV-binding form is of the common chromophore-binding structure seen for BphPs, although it may also facilitate PCB-binding (Fig. 1).
Dark and photoactive light conversion of Pfr and Pr BV-AM1_5894ΔHK (a,c) and Pfr and Pr PCB-AM1_5894ΔHK (b,d). Upper panels show Pfr to Pr photoconversion of the Pfr saturated forms of (a) BV- and (b) PCB-AM1_5894ΔHK on exposure to darkness or saturating light emitting diodes (LEDs) at the indicated wavelength for the indicated length of time. Lower panels show Pr to Pfr photoconversion of the Pr saturated forms of (c) BV- and (d) PCB-AM1_5894ΔHK on exposure to saturating light at the indicated wavelength for the indicated time. The extent of photoconversion is shown as the change in absorbance at the Pr λmax, longer bars indicate higher conversion. Difference spectra insets show an example of each photoconversion.
The switching of PCB-AM1_5894ΔHK from its Pr to its Pfr state was more sensitive to light wavelength than BV-AM1_5894ΔHK. For both, 665 nm light was most efficient at inducing Pr to switch to Pfr (Fig. 6c,d). However, 710 nm and 638 nm incident light were similarly effective for BV-AM1_5894ΔHK (Fig. 6c). Unexpectedly, 450 nm light partially switches BV-AM1_5894ΔHK from Pr to Pfr states, as illustrated by both this switching and the slight reduction in the Pfr to Pr switching when comparing 450 nm-exposed BV-AM1_5894ΔHK to that left in the dark (Fig. 6a,c).
It is known that a conserved Cys residue in the PAS domain of BphPs (Cys11 for AM1_5894ΔHK, Fig. 1) is the position for covalent binding of BV5. To determine whether Cys11 was involved in binding PCB, as well as BV, we constructed a C11S-AM1_5894ΔHK mutant, then BV or PCB was added to lysed E. coli containing overexpressed C11S-AM1_5894∆HK before His-tag purification. Under non-denaturing conditions, both BV and PCB seemed to elute with protein together during size exclusion chromatographic purification. However, the BV-associated complex appears much weaker than for WT AM1_5894ΔHK with a very low chromophore ratio (Fig. 7c). Although there were protein/chromophore complexes with a similar retention time to that of the WT for PCB, the spectrum of PCB-C11S-AM1_5894ΔHK was different, with an extra peak at around 555 nm, likely to be “unspecified” protein-chromophore complexes (Fig. 7b). However, no Zn-induced fluorescence was observed on the SDS-polyacrylamide gel for the C11S-AM1_5894ΔHK mutant incubated with either BV or PCB (Fig. 7a,b). Thus, our data demonstrate that Cys11 of AM1_5894 is essential for covalent binding of both BV and PCB. Both chromophores showed non-covalently associated with the C11S-AM1_5894ΔHK mutant (Fig. 7b,c), but this association was unstable. We were unable to detect any photoconversion in this mutant, associated with either BV or PCB (Figure S4).
(a) SDS polyacrylamide gel analysis on reconstituted protein complexes. 1, wt:BV-AM1_5894ΔHK; 2, C11S:BV-C11S-AM1_5894ΔHK; 3, wt:PCB-AM1_5894ΔHK; 4, C11S:PCB-C11S-AM1_5894ΔHK; 5 and 6, the purified AM1_5894ΔHKC11S protein before and after incubation in the absence of BV or PCB, respectively. Covalently bound chromophore was detected by Zn induced fluorescence (Zn). CBB, Coomassie brilliant blue. (b) The online spectral comparison of PCB-AM1_5894ΔHK with PCB-C11S-AM1_5894ΔHK after BioSec2000 purification; (c) the online spectral comparison of BV-AM1_5894ΔHK with BV-C11S-AM1_5894ΔHK after BioSec2000 purification.
Discussion
Acaryochloris uses red-shifted Chl d to capture light for photosynthesis in red/far-red light enriched environments22. To optimize the efficiency of light capture, Acaryochloris dynamically regulates chlorophyll- and phycobiliprotein-based light-harvesting complexes in a manner dependent upon light intensity and quality30,36. Here, we characterized the spectral properties of AM1_5894, a newly described BphP in Acaryochloris. The domain architecture of AM1_5894 resembles that of other canonical BphPs, having an N-terminal photosensory core module of PAS-GAF-PHY and a C-terminal signal transduction domain (Fig. 1). However, GAF/PHY domains are phylogenetically similar to Bathy BphPs from soil dwelling bacteria (Figure S1). Almost all these Bathy BphPs have a Pfr ground state, suggesting that these soil dwelling diazotrophs use these BphPs for a non-standard response to the perception of red light10. Although AM1_5894 appears related to Bathy Bphs, it has a photocycle that resembles the majority of characterized phytochromes, having a Pr ground state that switches to the Pfr light-activated state when exposed to red light. AM1_5894 and Bathy BphPs also share the same signal output domain structure, with an HWE-type histidine kinase and a C-terminal receiver domain (Fig. 1). These characteristics place AM1_5894 in a cluster of BphPs termed type 2 (Figure S1)10 and as only type 2 BphPs share an HWE-type histidine kinase domain they are thought to share a common evolutionary origin, which is consistent with the hypothesis presented in Buchberger & Lamparter41. Questions clearly remain as to the origin of AM1_5894 in the Acaryochloris genome, as well as the similar BphP identified in Leptolyngbya PCC 7375.
AM1_5894 autocatalytically reconstitutes in vitro from the apoprotein and the chromophores BV and PCB. These holoproteins exist in two photoconvertible forms: a Pr form (dark-adapted; λPCB max[Pr] 682 nm, λBV max[Pr] 705 nm) and a Pfr form (light-activated state; λPCB max[Pfr] 734 nm, λBV max[Pfr] 758 nm). It is noted that λPCB max[Pr] 682 nm of PCB-AM1_5894 is quite different from λPCB max[Pr] 655 nm of Cph1, which might be a result of the different binding site or the different binding properties12. The ability to use different chromophores of AM1_5894ΔHK in combination with recently identified CBCRs in Acaryochloris provides the molecular basis for Acaryochloris to thrive in different ecological niches, especially in diverse light environments.
Based on sequence alignments of the GAF and PHY domains of AM1_5894 with other bacterial and plant phytochromes, we initially hypothesized that AM1_5894 would covalently bind BV, but not necessarily PCB (Fig. 1). To our surprise, Zn-induced fluorescence on SDS-polyacrylamide gels suggests that AM1_5894 forms a covalent attachment to PCB as well as BV. Our experiments with a C11S-AM1_5894ΔHK mutant suggested that PCB is covalently bound to Cys 11 of AM1_5894, although the binding mechanism is unclear (Fig. 2). The GAF domains from two CBCRs from Acaryochloris have also been suggested to covalently bind PCB and BV based on Zn-induced fluorescence on SDS-polyacrylamide gels37,38,39. Other BphPs from non-photosynthetic bacteria, such as Deinococcus radiodurans, Pseudomonas syringae and P. aeruginosa, also bind PCB (and PΦB) covalently, as well as their natural chromophore BV37,38. But since they do not produce PCB or PΦB, the covalent binding of PCB to these Bphs is not physiologically relevant.
When bound to BV, their spectra and Pr/Pfr switching spectral properties resemble those of BphPs, under more red-shifted sensory light wavelengths. The reconstituted BV- AM1_5894ΔHK protein complexes have Pr/Pfr absorption maxima of 705 and 758 nm, respectively. These are ~20 nm-red-shifted compared to the PCB-AM1_5894ΔHK Pr/Pfr absorption maxima of 682 and 734 nm. The cBphP from Fremyella diplosiphone has similar Pr/Pfr spectral profiles, with a protein domain structure resembling that of AM1_5894, i.e., a Cys residue in its PAS domain and no canonical Cys residue in the GAF domain that covalently binds BV but not PCB39. Mutational analysis suggests that the BV-binding Cys residue in the PAS domain (C24) was not the site of attachment46. In fact, mutation of the His residue (H267) located adjacent to the canonical PCB/PΦB binding site markedly affected BV-binding and spectral properties5,47. These data contrast with mutational analysis of BphPs in Pseudomonas aeruginosa, where mutation of the Cys residue in the PAS domain prevents covalent attachment of BV, while mutation of the corresponding conserved His residue does not46. The non-covalent attachment of PCB maintains a more conjugated chromophore, which could explain the red-shifted spectral properties in PCB-binding in cBphPs from Fremyella diplosiphone, compared with other PCB-phytochrome holoproteins. However, the abolition of the covalent attachment of PCB to the C11SAM1_5894ΔHK mutant indicates that this cysteine residue is the site of the PCB covalent binding (Fig. 7).
Acaryochloris is able to dynamically regulate its light-harvesting mechanism in response to different light conditions, reducing the level of phycobiliproteins (which absorb light at around 640 nm) in response to high levels of white light or under far-red (720 nm) light30,36. Under such conditions, phycobiliproteins are either unnecessary for light capture or unable to capture far-red light, respectively. We have shown that BV-AM1_5894 can switch from its Pr ground state to its Pfr light-activated state using light sources between 625–708 nm. It efficiently switches back to its ground state using light sources between 730–780 nm. The light wavelength range between 710–730 nm is physiologically relevant to Acaryochloris, since Chl d-protein complexes absorb maximally within this range in vivo. The presence of PCB-AM1_5894 in addition to CBCRs found in Acaryochloris provide it with an effective method of measuring changes in the light spectrum relevant to its photosynthetic absorption range, allowing it to regulate target gene expression. The ability of AM1_5894 to covalently bind both BV and PCB in vitro and possibly in vivo (both being present in Acaryochloris) offers much broader light detection compared with other phytochromes, giving an adaptive advantage to Acaryochloris in its niche environments.
Extending the search for GAF domains beyond typical Cphs and BphPs in the Acaryochloris genome identified at least 11 genes that encode CBCR-like GAF domains. To date, GAF domains from AM1_1186, AM1_1870 and AM1_1557, have been characterized37,38,39. The GAF domain of AM1_1186 binds PCB and acts as a so-called dual-Cys CBCR with a red/blue light photocycle38. A GAF domain from AM1_1557 (GAF domain 2) and AM1_1870 (GAF domain 3) both covalently bind PCB and BV. For both AM1_1552g2 and AM1_1870g3 bound to PCB, a red-green photocycle was observed, however this shifted to a far red-orange photocycle when PCB was replaced with BV37,39. The BV-bound recombinants have red-shifted absorption peaks at 697 nm for BV-AM1_1557g239 and 706 nm for BV-AM1_1870g337. Although the spectral range in which CBCRs function is considerably greater than for other phytochromes, including BphPs, it is likely that the red/far-red photocycle of AM1_5894 best overlaps with the photosynthetically-relevant spectral range for Acaryochloris, making it a better candidate for regulating the photoacclimation exhibited by Acaryochloris.
Experimental Procedures
Expression and purification of recombinant AmrBphP
One canonical phytochrome, AM1_5894, was identified from the Acaryochloris marina MBIC11017 genome. The 2544 bp AM1_5894 open reading frame was amplified from genomic DNA of Acaryochloris using the following primers to allow In-Fusion® (Clontech, CA, USA) cloning (forward: 5′-CGCGCGGCAGCCATATGGAAATTAGAGAACTAGCGATTT-3′, reverse: 5′-GTTAGCAGCCGGATCCTTAGGCTAAGAGTTGATGAATG-3′). The underlined sequences are 5′ overhangs specific to the E. coli expression vector pET15b, which introduces a hexa-histidine tag to the expressed protein. A clone harbouring a full length AM1_5894 gene with a single amino acid substitution of Gly639 →Asp is designated as full-length AM1_5894D639G was obtained. To improve the protein overexpression yield, the 1524 bp N-terminal photosensory domain (AM1_5894ΔHK, lacking its histidine kinase and receiver domains) was amplified from Acaryochloris genomic DNA, using Phusion polymerase (New England Biolabs Ltd., UK) and the primer pair AmBph_ITA_FW (GCAGCGGCCTGGTGCCGCGCGGCAGCCATATGGAAATTAGAGAACTAGCGATTTCT) and AmBph_ITA_RV (CCAACTCAGCTTCCTTTCGGGCTTTGTTAGCAGCCGTTATTCCTGCTGCTGGTTTTCTCC). The underlined 3′ sequences correspond to pET15b sequences, which allowed assembly of the PCR product into pET15b, using a Gibson Assembly® master mix (New England Biolabs). A Cys11Ser mutant of AM1_5894ΔHK (C11S-AM1_5894ΔHK) was synthesized as a gBlock (Integrated DNA Technologies, Corlaville, IA, USA), with the overlapping pET15b sequence. It was also assembled into pET15b using a Gibson Assembly® master mix (New England Biolabs). The resulting three constructs were used to transform DH5α E. coli cells. The fidelity of all clones (including the point mutation that caused the Gly639Asp mutation in the full length clone) was confirmed before being used to transform Rosetta E. coli cells.
Rosetta E. coli (BL21, DE3) cultures harbouring the pET15b constructs were grown at 37 °C for 1 h and then at 21 °C until OD600 = 0.5 in LB media, supplemented with ampicillin and chloramphenicol at 100 μg/ml and 35 μg/ml, respectively. Expression was induced by the addition of 330 μM Isopropyl β-D-1-thiogalactopyranoside (IPTG) and cultures were grown at 10 °C for 48 h to limit aggregation of the expressed protein. The cells were harvested by centrifugation and the cell pellet resuspended in binding buffer (50 mM potassium phosphate buffer, pH 8.0, 10% (v/v) glycerol,) prior to cell lysis. Cells were lysed using a pre-chilled French Pressure Cell (GlenMills, NJ, USA) at approximately 1500 psi. His-tagged AM1_5894 protein was purified from the clarified crude protein extract using a His Gravitrap column (GE Healthcare, Sweden) and eluted with 500 mM imidazole. In some instances, BV or PCB was added to the crude protein extract prior to purification. Eluted protein was concentrated using a 3 kDa cut-off protein concentrator (Merck Millipore, MA, USA) and rinsed in 50 mM potassium phosphate buffer in order to dilute imidazole concentration to the final of <50 mM.
Chromophore reconstitution
Overexpressed AM1_5894ΔHK or full-length AM1_5894 (30 μM concentration of crude extract) was reconstituted in vitro with 100 μM of either BV (Frontier Scientific, USA) or PCB. PCB was isolated from powdered Spirulina (Lifestream International, NZ) by a modified methanolysis method48. PCB concentrations were determined spectroscopically using an extinction coefficient (ε690nm) of 37,900 M−1cm−1 in methanol/5%HCl49,50. For confirmation of covalent binding between the chromophore and AM1_5894 or AM1_5894ΔHK, reconstituted chromophore-protein complexes were analyzed by size exclusion chromatography using a BioSec2000 column (300 × 4.6 mm, Phenomenex) on a Shimadzu HPLC (25 mM phosphate buffer with pH 8.0 as the mobile phase, with a flow rate of 0.2 ml/min). The assembled BV-AM1_5894ΔHK or PCB-AM1_5894ΔHK was separated by SDS-polyacrylamide gel and Zn-induced fluorescence was monitored to detect protein-bound chromophores, after incubating the gel in 1 mM ZnCl2 for 10 min. Total protein composition for both was visualized with Coomassie Brilliant Blue staining.
Spectral characterization for photoconversion in vitro
All UV/visible absorption spectra were recorded with a UV2550 spectrophotometer (Shimadzu) at room temperature. For spectral analysis, approximately 30 μM His-tag purified AM1_5894ΔHK (as determined by its absorbance at 280 nm) was mixed with BV or PCB at a final concentration of 20 μM and the assembly was monitored by measuring the increase in absorbance at 705 nm or 682 nm, respectively. The kinetics of switching between the Pr (a red-light absorbing form) and Pfr (a far red-light absorbing form) states of BV-AM1_5894ΔHK or PCB-AM1_5894ΔHK were monitored over time by measuring spectral changes on exposure to either 665 nm (Pfr to Pr switch) or 777 nm (Pr to Pfr) monowavelength LEDs (Figure S3). All LED applied in this experiment are dual in-line LEDs (DIP LED) powered by 1.5 V DC and 10–20 mA. The light intensity of the 665 nm LED is in the range between 3 and 8 μmol photons m−2 s−1 monitored by LI-COR light meter.
The optimal photoactive wavelengths for AM1_5894 were examined by exposure of the Pfr AM1_5894ΔHK bound to BV or PCB to either darkness or a series of narrow bandwidth LEDs (450, 522, 710, 735 and 777 nm). To determine the optimal wavelengths for Pr to Pfr conversion, the Pr state of BV-AM1_5894ΔHK or PCB-AM1_5894ΔHK also was illuminated with a series of narrow bandwidth LEDs (450, 522, 597, 638, 665, 710, and 735 nm). Emission spectra for these LEDs are shown in Figure S3. The times of exposure for switching are based on the kinetics of switching, corresponding to just prior to ‘saturation’ of switching. To study reversion back to its initial state, purified AM1_5894ΔHK protein were exposed to either 665 nm or 777 nm light for at least 1 min, with reversion experiments conducted 5 min apart. BV-AM1_5894ΔHK reversion from its Pr to Pfr state was activated by exposures of at least 15 min.
Additional Information
How to cite this article: Loughlin, P. C. et al. Spectral properties of bacteriophytochrome AM1_5894 in the chlorophyll d-containing cyanobacterium Acaryochloris marina. Sci. Rep. 6, 27547; doi: 10.1038/srep27547 (2016).
References
Auldridge, M. E. & Forest, K. T. Bacterial phytochromes: More than meets the light. Crit. Rev. Biochem. Mol. Biol. 46, 67–88, doi: 10.3109/10409238.2010.546389 (2011).
Karniol, B., Wagner, J. R., Walker, J. M. & Vierstra, R. D. Phylogenetic analysis of the phytochrome superfamily reveals distinct microbial subfamilies of photoreceptors. Biochem. J. 392, 103–116, doi: 10.1042/bj20050826 (2005).
Montgomery, B. L. & Lagarias, J. C. Phytochrome ancestry: sensors of bilins and light. Trends Plant Sci. 7, 357–366, doi: 10.1016/s1360-1385(02)02304-x (2002).
Ikeuchi, M. & Ishizuka, T. Cyanobacteriochromes: a new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochemical & Photobiological Sciences 7, 1159–1167, doi: 10.1039/b802660m (2008).
Lamparter, T., Michael, N., Mittmann, F. & Esteban, B. Phytochrome from Agrobacterium tumefaciens has unusual spectral properties and reveals an N-terminal chromophore attachment site. Proc. Natl. Acad. Sci. USA 99, 11628–11633, doi: 10.1073/pnas.152263999 (2002).
Lagarias, J. C. & Rapoport, H. Chromopeptides from phytochrome-the structure and linkage of the Pr form of the phytochrome. J. Am. Chem. Soc. 102, 4821–4828, doi: 10.1021/ja00534a042 (1980).
Quest, B., Hubschmann, T., Sharda, S., de Marsac, N. T. & Gartner, W. Homologous expression of a bacterial phytochrome - The cyanobacterium Fremyella diplosiphon incorporates biliverdin as a genuine, functional chromophore. FEBS J 274, 2088–2098, doi: 10.1111/j.1742-4658.2007.05751.x (2007).
Wagner, J. R., Zhang, J. R., Brunzelle, J. S., Vierstra, R. D. & Forest, K. T. High resolution structure of Deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution. J. Biol. Chem. 282, 12298–12309, doi: 10.1074/jbc.M611824200 (2007).
Karniol, B. & Vierstra, R. D. The pair of bacteriophytochromes from Agrobacterium tumefaciens are histidine kinases with opposing photobiological properties. Proc. Natl. Acad. Sci. USA 100, 2807–2812, doi: 10.1073/pnas.0437914100 (2003).
Rottwinkel, G., Oberpichler, I. & Lamparter, T. Bathy Phytochromes in Rhizobial Soil Bacteria. J. Bacteriol. 192, 5124–5133, doi: 10.1128/jb.00672-10 (2010).
Bussell, A. N. & Kehoe, D. M. Control of a four-color sensing photoreceptor by a two-color sensing photoreceptor reveals complex light regulation in cyanobacteria. Proc. Natl. Acad. Sci. USA 110, 12834–12839, doi: 10.1073/pnas.1303371110 (2013).
Ulijasz, A. T. et al. Cyanochromes are blue/green light photoreversible photoreceptors defined by a stable double cysteine linkage to a phycoviolobilin-type chromophore. J. Biol. Chem. 284, 29757–29772, doi: 10.1074/jbc.M109.038513 (2009).
Ulijasz, A. T. & Vierstra, R. D. Phytochrome structure and photochemistry: recent advances toward a complete molecular picture. Curr. Opin. Plant Biol. 14, 498–506, doi: 10.1016/j.pbi.2011.06.002 (2011).
Blankenship, R. E. & Chen, M. Spectral expansion and antenna reduction can enhance photosynthesis for energy production. Curr. Opin. Chem. Biol. 17, 457–461, doi: 10.1016/j.cbpa.2013.03.031 (2013).
Rockwell, N. C. et al. Eukaryotic algal phytochromes span the visible spectrum. Proc. Natl. Acad. Sci. USA 111, 3871–3876, doi: 10.1073/pnas.1401871111 (2014).
Hu, Q. et al. A photosystem I reaction center driven by chlorophyll d in oxygenic photosynthesis. Proc. Natl. Acad. Sci. USA 95, 13319–13323, doi: 10.1073/pnas.95.22.13319 (1998).
Tomo, T. et al. Identification of the special pair of photosystem II in a chlorophyll d-dominated cyanobacterium. Proc. Natl. Acad. Sci. USA 104, 7283–7288, doi: 10.1073/pnas.0701847104 (2007).
Airs, R. L. et al. Chlorophyll f and chlorophyll d are produced in the cyanobacterium Chlorogloeopsis fritschii when cultured under natural light and near-infrared radiation. FEBS Lett. 588, 3770–3777, doi: 10.1016/j.febslet.2014.08.026 (2014).
Gan, F. et al. Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light. Science 345, 1312–1317, doi: 10.1126/science.1256963 (2014).
Miyashita, H. et al. Chlorophyll d as a major pigment. Nature 383, 402–402, doi: 10.1038/383402a0 (1996).
Behrendt, L. et al. Endolithic chlorophyll d-containing phototrophs. ISME Journal 5, 1072–1076, doi: 10.1038/ismej.2010.195 (2011).
Kuhl, M., Chen, M., Ralph, P. J., Schreiber, U. & Larkum, A. W. D. A niche for cyanobacteria containing chlorophyll d . Nature 433, 820–820, doi: 10.1038/433820a (2005).
Larkum, A. W. D. et al. A Novel Epiphytic Chlorophyll d-containing Cyanobacterium Isolated from a Mangrove-associated Red Alga. J. Phycol. 48, 1320–1327, doi: 10.1111/j.1529-8817.2012.01233.x (2012).
Lopez-Legentil, S., Song, B., Bosch, M., Pawlik, J. R. & Turon, X. Cyanobacterial diversity and a new Acaryochloris-like symbiont from Bahamian Sea-squirts. PLoS One 6, e23938 doi: 10.1371/journal.pone.0023938 (2011).
Martinez-Garcia, M., Koblizek, M., Lopez-Legentil, S. & Anton, J. Epibiosis of oxygenic phototrophs containing chlorophylls a, b, c, and d on the colonial ascidian cystodytes dellechiajei. Microb. Ecol. 61, 13–19, doi: 10.1007/s00248-010-9694-6 (2011).
Miller, S. R. et al. Discovery of a free-living chlorophyll d-producing cyanobacterium with a hybrid proteobacterial/cyanobacterial small-subunit rRNA gene. Proc. Natl. Acad. Sci. USA 102, 850–855, doi: 10.1073/pnas.0405667102 (2005).
Mohr, R. et al. A new chlorophyll d-containing cyanobacterium: evidence for niche adaptation in the genus Acaryochloris. ISME Journal 4, 1456–1469, doi: 10.1038/ismej.2010.67 (2010).
Murakami, A., Miyashita, H., Iseki, M., Adachi, K. & Mimuro, M. Chlorophyll d in an epiphytic cyanobacterium of red algae. Science 303, 1633–1633, doi: 10.1126/science.1095459 (2004).
Chen, M. & Blankenship, R. E. Expanding the solar spectrum used by photosynthesis. Trends Plant Sci. 16, 427–431, doi: 10.1016/j.tplants.2011.03.011 (2011).
Duxbury, Z., Schliep, M., Ritchie, R. J., Larkum, A. W. D. & Chen, M. Chromatic photoacclimation extends utilisable photosynthetically active radiation in the chlorophyll d-containing cyanobacterium, Acaryochloris marina . Photosynthesis Res. 101, 69–75, doi: 10.1007/s11120-009-9466-7 (2009).
Blankenship, R. E. & Hartman, H. The origin and evolution of oxygenic photosynthesis. Trends Biochem. Sci. 23, 94–97, doi: 10.1016/s0968-0004(98)01186-4 (1998).
Chen, M., Quinnell, R. G. & Larkum, A. W. D. The major light-harvesting pigment protein of Acaryochloris marina . FEBS Lett. 514, 149–152, doi: 10.1016/s0014-5793(02)02315-3 (2002).
Chen, M., Floetenmeyer, M. & Bibby, T. S. Supramolecular organization of phycobiliproteins in the chlorophyll d-containing cyanobacterium Acaryochloris marina. FEBS Lett. 583, 2535–2539, doi: 10.1016/j.febslet.2009.07.012 (2009).
Marquardt, J., Senger, H., Miyashita, H., Miyachi, S. & Morschel, E. Isolation and characterization of biliprotein aggregates from Acaryochloris marina, a Prochloron-like prokaryote containing mainly chlorophyll d. FEBS Lett. 410, 428–432, doi: 10.1016/s0014-5793(97)00631-5 (1997).
Chan, Y.-W. et al. Pigment composition and adaptation in free-living and symbiotic strains of Acaryochloris marina. FEMS Microbiol. Ecol. 61, 65–73, doi: 10.1111/j.1574-6941.2007.00320.x (2007).
Gloag, R. S., Ritchie, R. J., Chen, M., Larkum, A. W. D. & Quinnell, R. G. Chromatic photoacclimation, photosynthetic electron transport and oxygen evolution in the Chlorophyll d-containing oxyphotobacterium Acaryochloris marina. Biochimica Et Biophysica Acta-Bioenergetics 1767, 127–135, doi: 10.1016/j.bbabio.2006.11.014 (2007).
Narikawa, R., Fushimi, K., Win, N. N. & Ikeuchi, M. Red-shifted red/green-type cyanobacteriochrome AM1_1870g3 from the chlorophyll d-bearing cyanobacterium Acaryochloris marina . Biochem. Biophysio. Res. Com 461, 390–395 (2015).
Narikawa, R., Enomoto, G., Win, N. N., Fushimi, K. & Ikeuchi, M. A New Type of Dual-Cys Cyanobacteriochrome GAF Domain Found in Cyanobacterium Acaryochloris marina, which has an unusual Red/Blue Reversible Photoconversion Cycle. Biochemistry 53, 5051–5059, doi: 10.1021/bi500376b (2014).
Narikawa, R. et al. A biliverdin-binding cyanobacteriochrome from the chlorophyll d-bearing cyanobacterium Acaryochloris marina . Sci. Rep. 5, 7950, doi: 10.1038/srep07950 (2015).
Duxbury, Z., Willows, R. D., Smith, P. M. & Chen, M. In Photosynthesis Research for Food, Fuel and the Future (ed Kuang, T. Y., Lu, C. & Zhang, L. ) 638–641 (Zheijang University Press, Inc., 2013).
Buchberger, T. & Tilman Lamparter, T. Streptophyte phytochromes exhibit an N-terminus of cyanobacterial origin and a C-terminus of proteobacterial origin. BMC Res. Notes 8, 144, doi: 10.1186/s13104-015-1082-3 (2015).
Hill, C., Gartner, W., Towner, P., Braslavsky, S. E. & Schaffner, K. Expression of phytochrome apoprotein from Avena sativa in Escherichia coli and formation of photoactive chromoproteins by assembly with phycocyanobilin. Eur. J. Biochem. 223, 69–77, doi: 10.1111/j.1432-1033.1994.tb18967.x (1994).
Inomata, K. et al. Sterically locked synthetic bilin derivatives and phytochrome Agp1 from Agrobacterium tumefaciens form photoinsensitive Pr- and Pfr-like adducts. J. Biol. Chem. 280, 24491–24497, doi: 10.1074/jbc.M504710200 (2005).
van Thor, J. J. et al. Light-induced proton release and proton uptake reactions in the cyanobacterial phytochrome Cph1. Biochemistry 40, 11460–11471, doi: 10.1021/bi002651d (2001).
Yeh, K. C., Wu, S. H., Murphy, J. T. & Lagarias, J. C. Cyanobacterial phytochrome: implication for mechanism of phytochrome action. Plant Physiol. 114, 1464–1464 (1997).
Jorissen, H., Quest, B., Lindner, I., de Marsac, N. T. & Gartner, W. Phytochromes with noncovalently bound chromophores: The ability of apophytochromes to direct tetrapyrrole photoisomerization. Photochem. Photobiol. 75, 554–559, doi: 10.1562/0031-8655(2002)075<0554:pwnbct>2.0.co;2 (2002).
Davis, S. J., Vener, A. V. & Vierstra, R. D. Bacteriophytochromes: Phytochrome-like photoreceptors from nonphotosynthetic eubacteria. Science 286, 2517–2520, doi: 10.1126/science.286.5449.2517 (1999).
Storf, M. et al. Chromophore attachment to biliproteins: Specificity of PecE/PecF, a lyase-isomerase for the photoactive 31-Cys-α84-phycoviolobilin chromophore of phycoerythrocyanin. Biochemistry 40, 12444–12456, doi: 10.1021/bi010776s (2001).
Cole, W. J., Chapman, D. J. & Siegelman, H. W. Structure and properties of phycocyanobilin and related bilatrienes. Biochemistry 7, 2929–2935, doi: 10.1021/bi00848a033 (1968).
Arciero, D. M., Bryant, D. A. & Glazer, A. N. In vitro attachment of bilins to apophycocyanin. I. Specific covalent adduct formation at cysteinyl residues involved in phycocyanobilin binding in C-phycocyanin. J. Biol. Chem. 263, 18343–18349 (1988).
Acknowledgements
We would like to thank Dr. Artur Sawicki at Macquarie University for helpful comments and suggestions. MC holds an Australian Future Fellowship (FT120100464), with Australian Research Council financial support (DP120100286 and CE140100015).
Author information
Authors and Affiliations
Contributions
P.C.L., Z.D. and T.T.M.M. performed main experiments. P.C.L., P.M.C.S., R.D.W. and M.C. conceived the project and performed data analysis. P.C.L. and M.C. wrote the paper with input from other co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Loughlin, P., Duxbury, Z., Mugerwa, T. et al. Spectral properties of bacteriophytochrome AM1_5894 in the chlorophyll d-containing cyanobacterium Acaryochloris marina. Sci Rep 6, 27547 (2016). https://doi.org/10.1038/srep27547
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep27547
- Springer Nature Limited