Abstract
The (p)ppGpp signal molecules play a central role in the stringent response (SR) to adapt to nutrient starvation in bacteria, yet the carbohydrate starvation induced adaptive response and the roles of SR in this response is not well characterized, especially in Gram-positives. Here, two (p)ppGpp synthetases RelA and RelQ are identified in Streptococcus suis, an important emerging zoonotic Gram-positive bacterium, while only RelA is functional under glucose starvation. To characterize the roles of RelA/(p)ppGpp in glucose starvation response in S. suis, the growth curves and transcriptional profiles were compared between the mutant strain ΔrelA [a (p)ppGpp0 strain under glucose starvation] and its parental strain SC-19 [(p)ppGpp+]. The results showed great difference between SC-19 and ΔrelA on adaptive responses when suffering glucose starvation and demonstrated that RelA/(p)ppGpp plays important roles in adaptation to glucose starvation. Besides the classic SR including inhibition of growth and related macromolecular synthesis, the extended adaptive response also includes inhibited glycolysis and carbon catabolite repression (CCR)-mediated carbohydrate-dependent metabolic switches. Collectively, the pheno- and genotypic characterization of the glucose starvation induced adaptive response in S. suis makes a great contribution to understanding better the mechanism of SR.
Similar content being viewed by others
Introduction
In mammalian hosts, bacterial pathogens suffer various challenges in environmental stress and nutrient insufficiency, for example, high temperature, acidic environment, ROS (Reactive oxygen species) stimulation, lack of amino acids, metal ions or carbon sources and so on1,2,3,4,5,6. However, bacteria have evolved efficient stress response mechanisms to adapt to challenging environments. Among these responses, a special class of adaptive response induced by (p)ppGpp is called “stringent response (SR)”7. A wide array of physiological aspects, such as long-term persistence, virulence, biofilm formation and quorum sensing, have been reported to be affected by (p)ppGpp8,9,10,11.
The (p)ppGpp is synthesized by RelA/SpoT homologous proteins (RSH) through transferring a pyrophosphate moiety from ATP to GDP/GTP12,13,14. In Escherichia coli (E. coli) and some other Gram-negative bacteria, two RSH enzymes are involved in (p)ppGpp synthesis. The first enzyme RelA is recognized to respond to amino acid starvation in E. coli. The synthetic activity of RelA is activated by sensing ribosome idling reaction and then (p)ppGpp accumulation leads to reprogramming transcription, decreasing synthesis of stable RNAs and ribosome proteins and increasing amino acid biosynthesis15,16. Another homologous enzyme SpoT senses many other environmental stresses, such as starvation of carbon, iron, phosphate and fatty acids7,17,18. Different from E. coli, Gram-negative α-proteobacteria and Gram-positives contain a single RSH, such as RelBsu in Bacillus subtilis19, RelMtb in Mycobacterium tuberculosis8 and RelSmu in Streptococcus mutans20. Additionally, some small proteins with only (p)ppGpp synthetic activity are found in Firmicutes, such as B. subtilis and S. mutans19,20. Similar to RelA in E. coli, (p)ppGpp synthetic activity of RSH in Gram-positives can be also activated by interacting with idling ribosomes during amino acid starvation12. However, the mechanisms of SR induced by various stresses are different but not well characterized7. This includes the SR induced by carbon starvation.
Carbon resources are essential for all organisms. Carbon starvation causes a series of stress responses in bacteria, including the general stress response controlled primarily by carbon catabolite repression (CCR) and (p)ppGpp-mediated SR21,22,23. In E. coli, the genes regulated by regulators Crp and RpoS in CCR are RelA-dependent, implying that (p)ppGpp is at the apex of global regulation in carbon starvation24. However, the SR induced by carbon starvation is not yet fully characterized. The signaling pathway and global regulation during this process is largely unknown. About ten years ago, Battesti and Bouveret found that SpoT of E. coli can interact with the acyl carrier protein (ACP), the central cofactor of fatty acid synthesis. This interaction involves sensing the signals of fatty acid starvation and triggering SpoT-dependent (p)ppGpp accumulation25. Given that carbon deprivation can lead to fatty acid starvation through shrinkage of the acetyl-CoA pool produced during glycolysis26, fatty acid metabolism could also be the relay for carbon starvation, another nutritional stress sensed by SpoT7,25. This opens the first window to understand how the RSH or SR is activated by carbon starvation. Three years later, the same research group demonstrated that the interaction between RSH and ACP occurs also in Pseudomonas aeruginosa, but not in the Gram-positive B. subtilis and Streptococcus pneumoniae27. These preliminary studies indicate that the regulatory mechanism of carbon starvation induced SR varies among different species of bacteria, while the analogous mechanisms in Gram-positives need to be characterized.
Streptococcus suis is a very important Gram-positive bacterium that causes deadly infections in pigs and humans. As an emerging zoonotic pathogen, S. suis serotype 2 has become the predominant causative agent of adult human meningitis in Vietnam and Hong Kong28. Two large outbreaks of human infections were reported in China in 1998 and 2005, resulting in 229 infections and 52 deaths28,29. As a bacterial pathogen, host adaptation is one of the most important steps for pathogenesis. The relA gene had been found to be up-regulated during iron starvation in our previous study5, suggesting that RelA may play an important role in the adaptive response to nutrient starvation in S. suis. Investigation of the mechanism of S. suis adaptation to environments will contribute to understanding the transmission and pathogenesis of this important zoonotic pathogen. Concerning that the regulatory behaviour and mechanism of (p)ppGpp synthetases on adaptive response to carbon starvation may be different in Gram positive bacteria from Gram negatives27 and remain unraveled, this study aims to characterize the (p)ppGpp synthetases and their regulation on adaptive response to carbon starvation in S. suis.
Results
Identification of (p)ppGpp synthetases in S. suis
SR is an important adaptive response in bacteria, but until now, (p)ppGpp synthetase has not been characterized in S. suis. Searching the S. suis 05ZYH33 genome, we identified a RSH protein encoded by SSU05_2094 that consists of 733 amino acids. The N-terminal part is the catalytic domain for both hydrolysis and synthesis of (p)ppGpp and the C-terminus is recognized as the regulatory domain7. This protein was named RelA. Further amino acids sequence analysis showed that RelA of S. suis contains a RXKD motif (Fig. 1a) and shows higher similarity with SpoT rather than RelA of E. coli and other strains which contain two RSH in the genome (Fig. 2a)30,31. Additionally, a paralogous protein HP1060 encoded by SSU05_1060, henceforth designated RelQ, was found. It only contains a putative (p)ppGpp synthetase domain and lacks the hydrolase domain and regulatory domain (Fig. 1a), amino acids sequence analysis showed that the RelQ in S. suis had 78% identity with the RelQ in S. mutans. The phylogenetic tree revealed two clusters of the identified small alarmone synthetases (SASs) in Firmicutes and RelQ was clustered in the clade I group (Fig. 2b). To verify the (p)ppGpp synthetase activity of RelA and RelQ in vitro, soluble His-tagged recombinant RelA and RelQ were expressed in E. coli, purified (Fig. 1b) and then assayed for (p)ppGpp synthetase activity with [γ-32P]-ATP in the presence of 2 mM ATP and either 1.3 mM GTP or 1.3 mM GDP. The generated (p)ppGpp was detected by thin layer chromatography (TLC). The results showed that both ppGpp and pppGpp could be synthesized from GDP and GTP respectively by His-RelQ in vitro. Unexpectedly, ppGpp was detected when GTP was the substrate. It was a possible contaminant in the GTP that might arise from the intrinsic hydrolysis of GTP. In contrast, pppGpp could also be synthesized from GTP by His-RelA, but ppGpp was not detected when GDP was used as the substrate of His-RelA (Fig. 1c). The (p)ppGpp hydrolase activity of RelA and RelQ were further tested by detecting the hydrolytic product pyrophosphate (PPi) in vitro (Fig. 1d). There was no significant difference on the levels of PPi in the RelQ reaction (0.17 ± 0.02 μM) and negative control (0.19 ± 0.019 μM, no protein was added) (p > 0.05). We inferred that the small quantity of PPi came from the intrinsic hydrolysis of (p)ppGpp during storage or experiments. In contrast, approximately 1.57 μM PPi was detected in the RelA-catalyzed hydrolysis reaction. These results demonstrated that RelA is a bifunctional enzyme, which could synthesize and hydrolyze (p)ppGpp, whereas RelQ is a monofunctional (p)ppGpp synthetase in S. suis.
Expression and activity analysis of RelA and RelQ proteins of S. suis.
(a) Domain structures of RelA and RelQ of S. suis. The RXKD motif was labeled in the box, TGS, conserved domain on RSH C-terminal domain for uncharged ACP binding in E. coli, ACT, a conserved regulatory domain on RSH C-terminal domain. (b) SDS-PAGE analysis of purified recombinant RelA and RelQ proteins. (c) Thin-layer chromatography (TLC) analysis of the (p)ppGpp synthesis activity of the recombinant RelA and RelQ. Purified RelA or RelQ protein is assayed for (p)ppGpp synthetase activity in the presence of [γ-P32]-ATP, 2 mM ATP and either 1.3 mM GTP or GDP. Reaction mixtures were analyzed by TLC and autoradiography as described in Materials and Methods. (d) Hydrolase activity assays of recombinant RelA and RelQ by measuring the ppGpp hydrolysis product, PPi.
(p)ppGpp accumulation by S. suis strains during glucose starvation
To further verify the function of RelA and RelQ during glucose starvation, the mutant strains ΔrelA, ΔrelQ and ΔrelAΔrelQ were constructed using homologous recombination method and verified by PCR, RT-PCR and Southern blot. The (p)ppGpp accumulation in SC-19 and the three mutant strains during glucose starvation was studied by TLC analysis. When cells were cultured in the complete CDM, no (p)ppGpp was detected in all tested strains. In contrast, when cultured in the glucose-deficient CDM, (p)ppGpp synthesis could be detected in the relA-positive strains (SC-19 and ΔrelQ), but not relA-deletion strains (ΔrelA and ΔrelAΔrelQ) (Fig. 3). We further detected the ppGpp contents by anion exchange high performance liquid chromatography (HPLC) at the time point of 5 h, the sample collection point for microarray analysis (Fig. 4). When cultured in CDM containing 0.2% glucose, 119 ± 15 and 105 ± 19 pmoles/ml ppGpp were detected in SC-19 and ΔrelQ respectively, while no ppGpp was detected in relA-deletion strains. No ppGpp was detected in all the strains in CDM containing 1% glucose. These results indicated that glucose starvation can cause (p)ppGpp accumulation in S. suis, wherein RelA is the sole functional (p)ppGpp synthetase during glucose starvation. Whereas, although RT-PCR test have proved that relQ gene was expressed under glucose starvation, RelQ protein did not synthesize (p)ppGpp in ΔrelA under this condition.
The (p)ppGpp production by S. suis SC-19 and its mutant strains during glucose starvation.
The strains SC-19, ΔrelA, ΔrelQ and ΔrelAΔrelQ were grown in the MOPS-CDM containing 150 μCi/ml H3[32P]O4 and 1.0% or 0.2% glucose at 37 °C for 30 min. The formic acid extracts of the cells were subjected to TLC analysis as described in Materials and Methods.
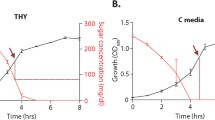
Growth of S. suis strains during glucose starvation
To investigate growth performance of the strains during carbon starvation, the wild type strain SC-19 and 3 mutant strains were grown in the complete CDM (containing 1% glucose) and glucose-deficient CDM (containing 0.2% glucose), respectively. In the complete CDM, RelQ inactivation did not affect the growth of S. suis, while the growth of RelA inactivated strains (ΔrelA and ΔrelAΔrelQ) slowed down when compared with SC-19 and ΔrelQ (Fig. 4a). In the glucose-deficient CDM, the growth rate of all strains was significantly decreased when compared with that in the complete CDM; RelA inactivated strains (ΔrelA and ΔrelAΔrelQ) showed a higher growth rate than SC-19 and ΔrelQ at the beginning of starvation and then more quickly turned into stationary phase; ΔrelQ displayed the similar growth performance as SC-19 (Fig. 4b). These results indicated that it is RelA but not RelQ contributing to the growth regulation of S. suis during glucose starvation.
Overview of microarray data
Growth curves and (p)ppGpp accumulation assays showed that RelA inactivation could influence S. suis growth and led to incapacity of (p)ppGpp synthesis during glucose starvation (Figs 3 and 4). To identify the roles of RelA/(p)ppGpp in global gene regulation in S. suis, we compared the transcriptional profiles of SC-19 [a (p)ppGpp+ strain] and ΔrelA [a (p)ppGpp0 strain during glucose starvation] in both glucose-abundant and -deficient CDM in early exponential phase by microarray analysis. We considered genes to be significantly induced or repressed if the absolute value of the expression ratio was > two-fold. qRT-PCR validation displayed the same trends observed in the microarrays (see Supplementary Tables S1 and S2). The results showed that 502 genes were up-regulated and 596 genes were down-regulated in SC-19, while 311 genes were up-regulated and 297 genes were down-regulated in ΔrelA during glucose starvation (see Fig. 5a and Supplementary Tables S3 and S4). More genes were found to be differentially expressed (DE) in SC-19 compared to ΔrelA during glucose starvation, suggesting a globally regulatory role of RelA/(p)ppGpp in S. suis under this condition. According to COG classification, the DE genes were classified into 19 categories (Fig. 5b). Among them were large numbers of genes involved in carbohydrate transport and metabolism, transcription and translation. Of note, plenty of genes in the glycolysis and extended metabolism pathways were differentially regulated between SC-19 and ΔrelA. In addition, many genes associated with cell cycle and cell wall/membrane biogenesis were down-regulated in SC-19, but not in ΔrelA. The details of these regulated genes are described below.
Summary of the differential expressed (DE) genes in S. suis SC-19 and ΔrelA response to glucose starvation (growth in 0.2% glucose vs 1% glucose).
(a) Venn diagrams show the up- (left) and down-regulated (right) genes. (b) The DE genes are classified into 19 functional categories. E, Amino acid transport and metabolism; G, Carbohydrate transport and metabolism; D, Cell cycle control, mitosis and meiosis; M, Cell wall/membrane biogenesis; H, Coenzyme transport and metabolism; V, Defense mechanisms; C, Energy production and conversion; P, Inorganic ion transport and metabolism; U, Intracellular trafficking and secretion; I, Lipid transport and metabolism; F, Nucleotide transport and metabolism; O, Posttranslational modification, protein turnover, chaperones; L, Replication, recombination and repair; T, Signal transduction mechanisms; Q, Secondary metabolites biosynthesis, transport and catabolism; K, Transcription; J, Translation; R, General function prediction only.
Typical (p)ppGpp-dependent SR
An important feature of classical SR is inhibition of biomacromolecule synthesis15. In this study, protein synthesis was inhibited by glucose starvation in SC-19. Compared to 26 DE genes in ΔrelA, more genes (69 genes) associated with protein translation were down-regulated in SC-19 (Fig. 6). Hereinto, 34 ribosomal protein genes were down-regulated in SC-19. The genes encoding transcription apparatus were also down-regulated by glucose starvation in SC-19, such as translation initiation factors IF-1 (SSU05_0092) and IF-3 (SSU05_1270), translation elongation factors EF-Tu (SSU05_0530), EF-Ts (SSU05_1979), EF-G (SSU05_0922) and EF-P (SSU05_1823, SSU05_1824). This is compatible with the proposed mechanism of direct down-regulation of rRNA synthesis during SR7. Different from in SC-19, the expression changes of the protein synthesis related genes in ΔrelA were not regular during glucose starvation. In ΔrelA, 11 genes encoding ribosomal proteins were down-regulated, while another 6 were up-regulated. At the same time, translation initiation factors and translation elongation factors was not obviously repressed except IF-3. These results suggested that RelA/(p)ppGpp inhibited the protein synthesis by down-regulating the expression of ribosomal proteins and translation factors during glucose starvation.
Heat maps of some important DE genes in the S. suis response to glucose starvation.
Heat maps of log 2 expression ratios for the SC-19 and ΔrelA for the ribosomal protein genes, sugar transporter, CCR controlled genes and genes related to transcription apparatus, DNA replication, cell cycle and cell wall biogenesis are shown.
Besides protein synthesis, DNA replication was also inhibited in SC-19 during glucose starvation. These down-regulated genes including: dnaB (SSU05_2158, SSU05_2159) encoding DNA helicase, ssb (SSU05_1833) encoding single-stranded DNA binding protein, dnaG (SSU05_1429) encoding primase, rnhB (SSU05_0996) encoding ribonuclease HII and four subunits of DNA polymerase III (SSU05_0662, SSU05_1540, SSU05_1627 and SSU05_1954). In contrast, only two subunits of DNA polymerase III (SSU05_1540, SSU05_1954) and ssb, which were related to DNA replication, were regulated in ΔrelA. These results suggested that RelA/(p)ppGpp inhibited the DNA replication by down-regulating the relative enzymes during glucose starvation.
Meanwhile, the expression of cell division and growth related genes were repressed in SC-19 during glucose starvation as well. Fourteen genes encoding essential proteins for cell division were down-regulated in SC-19, including ftsE (SSU05_1411), ftsI (SSU05_1354), ftsL (SSU05_1743), ftsW (SSU05_0526), ftsX (SSU05_1410), ftsZ (SSU05_0481), divIVA (SSU05_0487) and divIC (SSU05_0010). In contrast, only one gene encoding actin-like ATPase (SSU05_0479) involved in cell division was down-regulated in ΔrelA. These results suggested that RelA/(p)ppGpp inhibited the expression of cell division relative proteins during glucose starvation.
Finally, the expression of cell wall/membrane biogenesis genes was inhibited by glucose starvation in SC-19. These include the CPS biosynthesis gene cluster (cps2ABCDEFGHIJ, SSU05_0564-0573), genes encoding glycosyltransferases for cell wall biosynthesis (SSU05_1275, SSU05_1277 and SSU05_2144) and polysaccharide biosynthesis proteins (SSU05_1285, SSU05_1288). Comparing with SC-19, the expression of these genes was not obviously changed in ΔrelA. These results suggested that cps gene cluster was also inhibited by RelA/(p)ppGpp during glucose starvation.
Regulation on carbohydrate transport
The phosphoenolpyruvate-dependent phosphotransferase system (PTS) and ATP-binding cassette (ABC) transporters are two major mechanisms for carbohydrates uptake in bacteria22. As glucose is the first-line carbon source for most bacteria, glucose starvation caused obvious changes on expression of the genes involved in carbohydrate transport. In SC-19, the expression of 27 PTS components and 4 ABC-type sugar transporters were up-regulated. In contrast, only 13 PTS components were up-regulated while 2 PTS components down-regulated in ΔrelA. Besides lacEF (SSU05_1037-1038), transcription of 11 of the 13 up-regulated PTS components were also induced in SC-19. Two ABC sugar transporters which were highly expressed in SC-19 were also up-regulated in ΔrelA. Although many genes were up-regulated in both strains, the fold changes of most regulated sugar transport systems in SC-19 were greater than those in ΔrelA (Fig. 6). These results suggested that RelA/(p)ppGpp contributed to the expression of carbohydrate transporters during glucose starvation.
Regulation on glycolysis
The roles of RelA/(p)ppGpp in glycolysis and its interrelated pathways in S. suis, were interpreted using the microarray data in a central metabolic context. Gene expression ratios were overlaid onto central carbon metabolic maps (Fig. 7). The expression levels of genes between SC-19 and ΔrelA in glycolysis showed obvious difference. In glycolytic pathway, fbaABCD (SSU05_0336-0339), tpiA (SSU05_0531), gapA (SSU05_0155), gpmAB (SSU05_1638, SSU05_0520) and eno (SSU05_1503) encoding five reversible enzymes, which could convert fructose-1,6-bis-P to phosphoenol-pyruvate, were more than two-fold down-regulated in SC-19 during glucose starvation. Because gluconeogenesis is incomplete in S. suis, the main metabolic direction is from glucose to pyruvate32. More to the point, pfkA (SSU05_0543) and pykA (SSU05_0544), encoding 6-phosphofructokinase and pyruvate kinase respectively, which catalyzed two irreversible steps in glycolysis, were down-regulated in SC-19. In contrast, the expression level of genes in glycolysis did not change up to two fold in ΔrelA during glucose starvation. These results implied that glycolysis was repressed in S. suis by RelA/(p)ppGpp during glucose starvation (Fig. 7c).
Regulation of glucose starvation on glycolysis and carbohydrate utilization in S. suis SC-19 (a), ΔrelA (b) and the summary of SC-19 vs ΔrelA (c). Transcriptome data is overlaid on the selected metabolic pathways. Genes up-regulated > two-fold are shown in red, while genes down-regulated > two-fold are shown in blue. Genes whose expression is not changed up to two-fold are shown in black. The cre-box containing genes are marked by underline.
RelA/(p)ppGpp influences carbon catabolite repression
Carbon catabolite repression (CCR) is the most important regulation mechanism in bacteria when suffering carbon starvation33. Therein, catabolite control protein A (CcpA) and transcriptional anti-terminator containing PTS-regulatory domain (PRD) are the key regulators22,34,35. In order to find the connection between SR and CCR during glucose starvation, we focused on the genes regulated by both RelA/(p)ppGpp and CCR. The cre-box, the target of the negative regulator CcpA, was scanned in S. suis genome using Virtual Footprint33. 48 and 26 DE genes/operons were found to contain a cre-box in their promoter regions in SC-19 and in ΔrelA, respectively. Most of these genes were related to utilization of sugar resources including galactose, glycogen, maltose and glycerol and some specific amino acids (Fig. 7). Some cre-dependent operons were only up-regulated in SC-19. For example, the operon galKT (SSU05_0360, SSU05_0361) encoding the enzymes for conversion of galactose to Glc-1-p, was more than 38-fold overexpressed in SC-19, suggesting that use of galactose during glucose starvation was very important in S. suis36. The glgABCD operon (SSU05_1013–1016) encoding the enzymes for glycogen biosynthesis37 was approximately 20-fold up-regulated in SC-19 during glucose starvation. Another set of regulated genes were up-regulated both in SC-19 and ΔrelA, but induction was maximized in SC-19. For example, malQ1 and malQ2 (SSU05_2131 and SSU05_2132) encoding the 4-α-glucanotransferase, were up-regulated in both SC-19 and ΔrelA and malP (SSU05_1444) encoding maltodextrinphosphorylase, was up-regulated only in SC-19. These enzymes can convert maltose to Glc-1-p and their high-level expression may assist S. suis in using maltose38. The dhaKL (SSU05_1957, SSU05_1958), which encode the dihydroxyacetone kinases involved in glycerol utilization, were more than 80-fold and 70-fold induced in SC-19 and ΔrelA respectively, suggesting glycerol was also an important carbon source for S. suis during glucose starvation. The genes encoding enzymes associated with alcohol utilization were also up-regulated, including adhA (SSU05_0279) and adhE (SSU05_0280). The arcABC operon (SSU05_0624, SSU05_0626, SSU05_0627) encoding the arginine deiminase system (ADS) was significantly up-regulated in both SC-19 and ΔrelA and the fold change of this operon in SC-19 was as twice as that in ΔrelA. ADS catalyzes the fermentative catabolism of arginine and generates an ATP per one arginine consumed39. ADS may be another important way for energy supply in S. suis during glucose starvation. These genes/operons were typically cre-dependent and regulated by CcpA. However, when analyzing the expression levels of these genes/operons, we found that RelA/(p)ppGpp was required for induction or maximal induction of them during glucose starvation (Fig. 6).
Some PTS operons, containing a transcriptional anti-terminator with a PTS-regulatory domain (PRD), are regulated by phosphorylating PRDs22. We found that some PRD-containing operons were significantly differently expressed between SC-19 and ΔrelA during glucose starvation, such as ulaAB and celABC which govern uptake of galactitol and cellobiose. The ulaAB operon (SSU05_0188 and SSU05_0189) carrying a PRD-containing transcriptional anti-terminator (SSU05_0187) was more than 46 fold up-regulated in SC-19, however, only less than 4 fold up-regulated in ΔrelA. Another transcriptional anti-terminator (SSU05_2076) containing PTS celABC (SSU05_2073–2075) was approximately 20 fold up-regulated in SC-19. These results showed that induction or maximal induction of these PRD-containing operons also required RelA/(p)ppGpp (Fig. 6).
Discussion
Environmental adaptation is an important issue for bacterial survival, growth and pathogenesis and SR is one of the most important mechanisms for environmental adaptation8,40. The signal molecules (p)ppGpp are the key player in SR induced by nutrient starvation14,16,41. In this study, two (p)ppGpp synthetases are identified in the emerging zoonotic pathogen S. suis. One is RelA containing all the four RSH domains and another is RelQ which only contains a (p)ppGpp synthetase domain (Fig. 1a). The relA gene was found to be up-regulated in S. suis during iron starvation in our previous study5, suggesting that relA may play a role in environmental adaptation and pathogenesis of S. suis. The synthetase and hydrolase activities of RelA were shown by the in vitro assays (Fig. 1), although RelA did not show the ability to synthesize ppGpp (Fig. 1c). Sajish et al. has reported that, bifunctional (synthesis and hydrolysis) RSH like SpoT in E. coli, contains a RXKD motif and prefer to utilize GTP, while monofunctional (synthesis) RSH like RelA in E. coli contains an EXDD motif and prefer to utilize GDP30,31. Sequence analysis showed that RelA in S. suis is closer to SpoT but not to RelA in E. coli and it has the typical characteristic in bifunctional RSH that contains a RXKD motif. That in S. suis the preference of RelA on GTP rather than GDP may be the reason why no ppGpp was detected in present of GDP. At the same time, we report for the first time RelQ and its (p)ppGpp synthetic activity in S. suis in vitro (Fig. 1c). Previous studies have identified several SASs which had the (p)ppGpp synthetic capacity in members of class Firmicutes, such as YjbM and YwaC in B. subtilis and RelP and RelQ in Streptococcus mutans19,20. The RelQ identified in S. suis had 78% identity of amino acid sequence with the RelQ in S. mutans, which could synthesize (p)ppGpp during amino acid starvation in S. mutans20, we infer that RelQ in S. suis is also functional during amino acid starvation. Unexpectedly in Fig. 1c, there was an excrescent point ppGpp when GTP was used as the substrate of RelQ. Recently, Anthony et al. reported that RelQEF synthesizes ppGpp more efficiently than pppGpp42, that may be the reason why ppGpp was detected largely in our test when possible GDP contamination existed in GTP. In Bacillus subtilis, initiative suppressor mutations in SASs could partially relieve the growth defect of relA mutant43. However, no initiative mutation was found in the relQ gene of S. suis relA mutant.
Carbohydrates are important nutrients for most bacteria and carbon starvation can induce SR in many species of bacteria, whereas its mechanisms are largely unclear, especially in Gram-positives24,27. To disclose the functional roles of the RSH proteins RelA and RelQ in regulation of SR in S. suis, we constructed three isogenic mutant strains ΔrelA, ΔrelQ and ΔrelAΔrelQ. Their (p)ppGpp accumulation during glucose starvation was detected by TLC and HPLC analysis. It is interesting that although relQ was expressed on transcriptional level in both SC-19 and ΔrelA during glucose starvation, (p)ppGpp can only be detected in the relA-positive strains (SC-19 and ΔrelQ) but not in relA-deleted strains (ΔrelA and ΔrelAΔrelQ) (Fig. 3), demonstrating that RelA was the sole functional (p)ppGpp synthetase in S. suis during glucose starvation (i.e. RelQ is not functional under this condition). Thus, under glucose starvation, ΔrelA can be considered as a (p)ppGpp0 strain of S. suis and was used to study the regulatory functions of (p)ppGpp.
To characterize the roles of RelA/(p)ppGpp in glucose-starvation induced adaptive response in S. suis, we compared and analyzed the glucose starvation responses of SC-19 and ΔrelA on growth curves and transcriptional profiles in this study. Comparing with SC-19, ΔrelA had a little lower growth rate when grown in the CDM containing 1% glucose (Fig. 4a). In contrast in the CDM containing 0.2% glucose, the growth of SC-19 lagged ΔrelA at the beginning of glucose starvation, but then ΔrelA declined more quickly (Fig. 4b). The different growth phenotypes between SC-19 and ΔrelA suggested that RelA/(p)ppGpp regulates the growth of S. suis during glucose starvation. These phenotypes were also reflected on the transcriptional levels of related genes from the results of microarray, including the typical SR on macromolecular synthesis and a series of special regulation on carbon metabolism. DNA replication, protein translation, cell division and cell wall/membrane biogenesis were significantly inhibited during glucose starvation in SC-19 (Fig. 6). These changes on macromolecular synthesis are the typical RelA/(p)ppGpp-mediated SR, which could be induced by amino acid starvation in many bacteria16,44,45. At the same time, glucose starvation induces a special regulation pattern on carbon metabolism in SC-19: all the glycolysis-related genes were down-regulated, while the CCR controlled genes were significantly activated by glucose starvation in SC-19 (Fig. 7a), suggesting that SC-19 has adjusted to reduce consumption of glucose and tries its best to uptake and utilize other carbon resources, such as galactose, maltose, glycerol and even amino acids like arginine (Fig. 6). In contrast, glucose starvation does not regulate the glycolysis-related genes in ΔrelA (Fig. 7b). It is well known that RelA/(p)ppGpp-mediated SR directly inhibit the macromolecular synthesis15, in this study we could conclude that, RelA/(p)ppGpp also plays important roles on the regulation of the glycolysis-related genes during glucose starvation adaptive response. Taken together, S. suis inhibits its macromolecular synthesis, cell cycle and adjusts its carbohydrate metabolisms to adapt to glucose starvation for long-term survival.
It is well known that CCR is a key player on regulation of the uptake and utilization of carbon resources32. In the present study, different transcriptional patterns of the CCR regulons were observed between SC-19 and ΔrelA under glucose starvation (Fig. 7), indicating an important role of RelA/(p)ppGpp in this process. For example, some CCR regulons such as glgABCD, galKT and malP were only up-regulated in SC-19 but not in ΔrelA, while other CCR regulons such as malQ, dhaKL and manB were induced at higher levels in SC-19 than in ΔrelA. This may be due to the different expressional levels of genes in glycolysis between SC-19 and ΔrelA. In glucose metabolism, the intermediates and their derivatives in glycolysis act as the indicators for CCR control, such as fructose-1,6-bisphosphate and glucose-6-phosphate22. During glucose starvation, the inhibited glycolysis in SC-19 leads to decreased concentration of the indicators and consequently results in significant derepression and up-regulation of the CCR regulons. In contrast, the transcriptional levels of the genes in glycolytic pathway do not changed in ΔrelA [(p)ppGpp0]. These findings indicate that an intersectional link should exist between the RelA/(p)ppGpp and CCR systems, at least in regulation of the carbon starvation induced SR.
Early studies have demonstrated that fatty acid (carbon) starvation is sensed by ACP/SpoT interaction in E. coli and the C-terminal TGS domain of SpoT is essential for this interaction25,27. However, our Bacterial Two-Hybrid analysis demonstrated that neither RelA nor RelQ can interact with ACP in S. suis (see Supplementary Fig. S1). Therefore, the signaling pathway of carbon starvation-induced SR in S. suis is different from that in E. coli. We infer that there might be other protein(s) other than ACP functioning as RSH-interacting partner(s) to transmit the carbon starvation signal in S. suis. Given that (p)ppGpp could not be synthesized in ΔrelA by RelQ during glucose starvation, we speculate that it might be due to absence of the carbon-starvation-sensing domain in RelQ. We currently are trying to identify the potential RelA-interacting partner(s) that can sense carbon starvation and induce stringent response in the zoonotic pathogen S. suis.
In conclusion, two (p)ppGpp synthetases RelA and RelQ are identified in S. suis, while only RelA is functional during glucose starvation. The RelA/(p)ppGpp-mediated stringent response plays important roles in adaptation to glucose starvation. Besides the classical SR including inhibition of growth and related macromolecular synthesis, the glucose-starvation induced adaptive response also includes inhibited glycolysis and extended CCR-mediated carbohydrate-dependent metabolic switches. This makes a great contribution to understanding better the mechanisms of carbon starvation induced stringent response in this important zoonotic pathogen.
Materials and Methods
Bacterial strains, plasmids and growth conditions
All the bacterial strains and plasmids used in this study are listed in Supplementary Table S5. S. suis serotype 2 strain SC-19 was isolated from a diseased pig in Sichuan province of China in 20054. S. suis was grown in tryptic soy broth (TSB) or on tryptic soy agar (TSA; Difco, France) plates containing 5% newborn bovine serum (Sijiqing, Hangzhou, China). E. coli DH5α was cultured in/on Luria-Bertani (LB) broth or plate (Oxoid, Basingstoke, UK). The chemically defined medium (CDM) for S. suis46 was modified when necessary (see Supplementary Table S6). For 32P-labeling in vivo, MOPS-CDM (the phosphate compounds-deleted CDM supplemented with 40 mM MOPS, 4 mM Tricine, 0.28 mM K2SO4 and 50 mM NaCl) was used. When necessary, antibiotics were added to the plate or broth at the following concentrations: 100 μg/ml spectinomycin (Spc), 2.5 μg/ml erythromycin (Erm) for S. suis; 50 μg/ml Spc, 180 μg/ml Erm, 50 μg/ml kanamycin (Kan) for E. coli.
Expression and purification of RelA and RelQ
The coding sequences of relA and relQ were amplified from the genomic DNA of S. suis SC-19 using primers relAF/relAR and relQF/relQR (see Supplementary Table S7), respectively. The primers were designed according to the sequences of genes SSU05_2094 and SSU05_1060 of S. suis 05ZYH33 (GenBank accession no. CP000407) and cloned into a prokaryotic expression vector pET-28a (Novagen, Shanghai, China), respectively. The resultant plasmids pET28a-relA and pET28a-relQ were confirmed by DNA sequencing and transformed into E. coli BL21(DE3) for expression of His-tagged recombinant proteins, respectively. The bacteria were induced by 1 mM isopropyl-beta-D-thiogalactopyranoside (IPTG) at 37 °C for 3 h. Purification of the recombinant proteins was achieved using Ni-NTA agarose (Bio-Rad, Shanghai, China) under native condition according to the manufacturer’s instructions. Electrophoresis was carried out with 12% SDS-PAGE.
(p)ppGpp synthetase/hydrolase activity assays
(p)ppGpp synthetase activity of RelA and RelQ assays was performed as described previously19. The reaction was carried out at a final volume of 25 μl containing 2 mM ATP, 10 μCi ml−1 [γ-P32]-ATP, 1.3 mM GTP or GDP and the purified 0.5 μg of recombinant RelA or RelQ in reaction buffer [50 mM Tris-acetate (pH 7.8), 3.3 mM magnesium acetate, 60 mM potassium acetate, 30 mM ammonium acetate, 1 mM dithiothreitol]. The mixture was incubated at 30 °C for 1 h and the reaction was stopped by adding 1 μl of 88% formic acid. 5 μl of each sample was spotted on polyethyleneimine cellulose plastic-backed Thin-Layer Chromatography (TLC) plate (Merck, Darmstadt Germany) and chromatographed in 1.5 M KH2PO4 (pH 3.4) for TLC analysis. TLC plates were exposed to a phosphor screen (GE Healthcare, NJ, USA) and radioactivity was scanned by Typhoon FLA 7000 IP (GE Healthcare).
The hydrolase activity assay was performed as described previously with some modification12. In brief, hydrolysis reaction mixtures contained 50 mM HEPES (pH 8.0), 150 mM NaCl, 1 mM DTT, 1.4 mM MnCl2, 0.7 mM ppGpp (TriLink, San Diego, USA) and 200 nM RelA or RelQ protein. In the negative control, the protein was replaced by ddH2O. After 5 min reaction at room temperature, the hydrolysis reaction product PPi was detected by measuring the fluorescent product (λex = 316/λem = 456 nm) proportional to the pyrophosphate present using Pyrophosphate Assay Kit according to the product manual (Sigma-Aldrich, St. Louis, USA).
Construction of mutant strains
To inactivate relA gene in S. suis strain SC-19, a thermosensitive homologous suicide vector pSET4s::relA carrying the left arm (L_arm, 731 bp), right arm (R_arm, 762 bp) and Erm resistance cassette (ermr) was constructed. The two arms were amplified from the chromosomal DNA of SC-19 using primers relAL01/relAL02 and relAR01/relAR02, respectively. The ermr was amplified from the plasmid pAT18 by using primers ermF/ermR (see Supplementary Table S7). The recombinant plasmid pSET4s::relA was electro-transformed into SC-19 and the strains were selected on Spc and Erm plates as described previously47. The suspected mutant strain ΔrelA was verified by PCR, RT-PCR and Southern blot analysis using standard protocols. The expression of genes upstream and downstream of relA was also tested by RT-PCR. Using similar methods, relQ gene was inactivated in strains SC-19 and ΔrelA, resulting in mutant strains ΔrelQ and ΔrelAΔrelQ.
Detection of intracellular (p)ppGpp
S. suis strains were grown in TSB with 5% newborn bovine serum as described above. At culture density of OD600nm ≈ 0.2, cells were collected by centrifugation at 3000 g for 2 min at room temperature and the pellets were washed and resuspended in MOPS-CDM to the same density. Then 20 μl of each cell suspension was added into the wells of 96-well microtiter plates and pre-warmed at 37 °C for 10 min. Glucose was the only carbon sources in the CDM, S. suis could not grow in the CDM without glucose. According to the previous report with some modification21, we used medium with 0.2% glucose to simulate glucose starvation. To simulate glucose starvation, cell suspensions in 0.2% glucose MOPS-CDM were mixed with 130 μl of 0.2% glucose MOPS-CDM containing 150 μCi/ml of H3[32P]O4. Cell suspensions mixed with 1% glucose MOPS-CDM containing 150 μCi/ml of H3[32P]O4 were used as controls. After incubation at 37 °C for 30 min, each 20 μl sample was removed and mixed with an equal volume of 13 M frozen formic acid, then the mixtures were refrozen and thawed twice. 5 μl of each thawed sample was analyzed by TLC as described above. The ppGpp contents of S. suis at the time point of sample collection of microarray analysis were further detected by anion exchange HPLC using a Mono Q 5/50 GL column (GE Healthcare) as previously described16. ppGpp standard was purchased from TriLink Biosciences (TriLink, San Diego, USA). Standard curves established that the linear range of detection of ppGpp is 50 nM–100 mM.
Growth in glucose starvation
The growth curves of S. suis WT and mutant strains during glucose starvation were tested as described below. Overnight cultured cells, including SC-19, ΔrelA, ΔrelQ and ΔrelAΔrelQ strains, were collected by centrifugation at 3000 g for 5 min and washed with CDM for three times. Then the pellets were resuspended in glucose starvation CDM (CDM containing 0.2% glucose) to the density of OD600 ≈ 0.1. Cells resuspended in complete CDM (CDM containing 1% glucose) were used as controls. All the samples were cultured at 37 °C and OD600nm value was measured once each hour.
Microarray analysis
To identify the genes regulated by RelA/(p)ppGpp during glucose starvation, microarray analysis was performed. Overnight cultured cells were washed with CDM for three times and resuspended in CDM containing 0.2% glucose and CDM containing 1% glucose to the density of OD600 ≈ 0.1 respectively. Cells in early exponential growth period (5 h in Fig. 4) were collected for microarray analysis. The test of each strain included three biological replicates. Briefly, total RNA were isolated and purified using QIAGEN RNeasy Mini Kit (Qiagen, Shanghai, China) according to the manufacturer’s instructions. 2 μg RNA was reverse-transcribed into cDNA and then the cDNA was transcribed into aaUTP labeled cRNA and purified. 4 μg cRNA was labeled with Cy3 NHS ester (GE healthcare) and purified. After fragmentation of the Cy3-cRNA, hybridization was performed using a Gene Expression Hybridization Kit (Agilent Technologies, Beijing, China). Finally, arrays were scanned by Agilent Microarray Scanner System with resolution of 5 μm. The signal intensities were normalized using Feature Extraction Software (Agilent Technologies) and transformed into log2 values. The microarray data have been submitted to the NCBI Gene Expression Omnibus (GEO) functional genomics data repository under the accession number GSE70092.
Quantitative RT-PCR (qRT-PCR)
Primers (see Supplementary Table S7) were designed based on the S. suis 05ZYH33 genome sequence. RNA extraction was carried out as described in microarray analysis. qRT-PCR was performed on an ABI 7300 HT Sequence Detection System using the ABI Power SYBE Green PCR Master Mix. The expression level of each tested gene was normalized to those of gapdh, which did not show any change in expression across all culture condition in both strains SC-19 and ΔrelA. Data were reported as mean relative expression levels (±standard deviation) under glucose starvation versus normal culture condition.
Statistical analysis
All the experiments were performed at least thrice with triplicate repeats. The means of two groups were compared using Student’s t test (unpaired, 2-tailed), with p < 0.05 considered to be statistically significant. Statistical analysis was performed using GraphPad Prism 6 (San Diego, USA). Microarray data were analyzed using GeneSpring Software 5.0 (Silicon Genetics, CA, USA). Those genes with greater than two-fold change ratios were regarded as differentially expressed genes.
Additional Information
How to cite this article: Zhang, T. et al. The roles of RelA/(p)ppGpp in glucose-starvation induced adaptive response in the zoonotic Streptococcus suis. Sci. Rep. 6, 27169; doi: 10.1038/srep27169 (2016).
References
Zeng, X. et al. Microarray analysis of temperature-induced transcriptome of Streptococcus suis serotype 2. Vector Borne Zoonotic Dis 11, 215–21 (2011).
Li, J. S., Bi, Y. T., Dong, C., Yang, J. F. & Liang, W. D. Transcriptome analysis of adaptive heat shock response of Streptococcus thermophilus. PLoS One 6, e25777 (2011).
Schlafer, S. et al. pH landscapes in a novel five-species model of early dental biofilm. PLoS One 6, e25299 (2011).
Zhang, T. et al. A Fur-like protein PerR regulates two oxidative stress response related operons dpr and metQIN in Streptococcus suis. BMC Microbiol 12, 85 (2012).
Li, W., Liu, L., Chen, H. & Zhou, R. Identification of Streptococcus suis genes preferentially expressed under iron starvation by selective capture of transcribed sequences. FEMS Microbiol Lett 292, 123–33 (2009).
Svensater, G., Bjornsson, O. & Hamilton, I. R. Effect of carbon starvation and proteolytic activity on stationary-phase acid tolerance of Streptococcus mutans. Microbiology 147, 2971–9 (2001).
Potrykus, K. & Cashel, M. (p)ppGpp: still magical? Annu Rev Microbiol 62, 35–51 (2008).
Dahl, J. L. et al. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci USA 100, 10026–31 (2003).
Geiger, T. et al. Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infect Immun 78, 1873–83 (2010).
Kazmierczak, K. M., Wayne, K. J., Rechtsteiner, A. & Winkler, M. E. Roles of rel(Spn) in stringent response, global regulation and virulence of serotype 2 Streptococcus pneumoniae D39. Mol Microbiol 72, 590–611 (2009).
Taylor, C. M. et al. Listeria monocytogenes relA and hpt mutants are impaired in surface-attached growth and virulence. J Bacteriol 184, 621–8 (2002).
Avarbock, D., Avarbock, A. & Rubin, H. Differential regulation of opposing RelMtb activities by the aminoacylation state of a tRNA.ribosome.mRNA.RelMtb complex. Biochemistry 39, 11640–8 (2000).
Cashel, M. Preparation of guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp) from Escherichia coli ribosomes. Anal Biochem 57, 100–7 (1974).
Cashel, M. Regulation of bacterial ppGpp and pppGpp. Annu Rev Microbiol 29, 301–18 (1975).
Dalebroux, Z. D. & Swanson, M. S. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol 10, 203–12 (2012).
Traxler, M. F. et al. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 68, 1128–48 (2008).
Seyfzadeh, M., Keener, J. & Nomura, M. spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc Natl Acad Sci. USA 90, 11004–8 (1993).
Vinella, D., Albrecht, C., Cashel, M. & D’Ari, R. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol Microbiol 56, 958–70 (2005).
Nanamiya, H. et al. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol Microbiol 67, 291–304 (2008).
Lemos, J. A., Lin, V. K., Nascimento, M. M., Abranches, J. & Burne, R. A. Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol Microbiol 65, 1568–81 (2007).
Ojha, A. K., Mukherjee, T. K. & Chatterji, D. High intracellular level of guanosine tetraphosphate in Mycobacterium smegmatis changes the morphology of the bacterium. Infect Immun 68, 4084–91 (2000).
Gorke, B. & Stulke, J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6, 613–24 (2008).
Song, M. et al. ppGpp-dependent stationary phase induction of genes on Salmonella pathogenicity island 1. J Biol Chem 279, 34183–90 (2004).
Traxler, M. F., Chang, D. E. & Conway, T. Guanosine 3′,5′-bispyrophosphate coordinates global gene expression during glucose-lactose diauxie in Escherichia coli. Proc Natl Acad Sci USA 103, 2374–9 (2006).
Battesti, A. & Bouveret, E. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol Microbiol 62, 1048–63 (2006).
DiRusso, C. C. & Nystrom, T. The fats of Escherichia coli during infancy and old age: regulation by global regulators, alarmones and lipid intermediates. Mol Microbiol 27, 1–8 (1998).
Battesti, A. & Bouveret, E. Bacteria possessing two RelA/SpoT-like proteins have evolved a specific stringent response involving the acyl carrier protein-SpoT interaction. J Bacteriol 191, 616–24 (2009).
Gottschalk, M., Xu, J., Calzas, C. & Segura, M. Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? Future Microbiol 5, 371–91 (2010).
Feng, Y., Zhang, H., Ma, Y. & Gao, G. F. Uncovering newly emerging variants of Streptococcus suis, an important zoonotic agent. Trends Microbiol 18, 124–31 (2010).
Sajish, M., Kalayil, S., Verma, S. K., Nandicoori, V. K. & Prakash, B. The significance of EXDD and RXKD motif conservation in Rel proteins. J Biol Chem 284, 9115–23 (2009).
Sajish, M., Tiwari, D., Rananaware, D., Nandicoori, V. K. & Prakash, B. A charge reversal differentiates (p)ppGpp synthesis by monofunctional and bifunctional Rel proteins. J Biol Chem 282, 34977–83 (2007).
Willenborg, J., de Greeff, A., Jarek, M., Valentin-Weigand, P. & Goethe, R. The CcpA regulon of Streptococcus suis reveals novel insights into the regulation of the streptococcal central carbon metabolism by binding of CcpA to two distinct binding motifs. Mol Microbiol 92, 61–83 (2014).
Iyer, R., Baliga, N. S. & Camilli, A. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J Bacteriol 187, 8340–9 (2005).
Almengor, A. C., Kinkel, T. L., Day, S. J. & McIver, K. S. The catabolite control protein CcpA binds to Pmga and influences expression of the virulence regulator Mga in the Group A streptococcus. J Bacteriol 189, 8405–16 (2007).
Stulke, J., Arnaud, M., Rapoport, G. & Martin-Verstraete, I. PRD–a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol Microbiol 28, 865–74 (1998).
Cai, J., Tong, H., Qi, F. & Dong, X. CcpA-dependent carbohydrate catabolite repression regulates galactose metabolism in Streptococcus oligofermentans. J Bacteriol 194, 3824–32 (2012).
Ugalde, J. E. et al. Gene organization and transcription analysis of the Agrobacterium tumefaciens glycogen (glg) operon: two transcripts for the single phosphoglucomutase gene. J Bacteriol 180, 6557–64 (1998).
Park, J. T. et al. Role of maltose enzymes in glycogen synthesis by Escherichia coli. J Bacteriol 193, 2517–26 (2011).
Gruening, P., Fulde, M., Valentin-Weigand, P. & Goethe, R. Structure, regulation and putative function of the arginine deiminase system of Streptococcus suis. J Bacteriol 188, 361–9 (2006).
Jain, V., Kumar, M. & Chatterji, D. ppGpp: stringent response and survival. J Microbiol 44, 1–10 (2006).
Wang, J. D., Sanders, G. M. & Grossman, A. D. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell 128, 865–75 (2007).
Gaca, A. O. et al. From (p)ppGpp to (pp)pGpp: Characterization of Regulatory Effects of pGpp Synthesized by the Small Alarmone Synthetase of Enterococcus faecalis. J Bacteriol 197, 2908–19 (2015).
Srivatsan, A. et al. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet 4, e1000139 (2008).
Durfee, T., Hansen, A. M., Zhi, H., Blattner, F. R. & Jin, D. J. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol 190, 1084–96 (2008).
Ferullo, D. J. & Lovett, S. T. The stringent response and cell cycle arrest in Escherichia coli. PLoS Genet 4, e1000300 (2008).
van de Rijn, I. & Kessler, R. E. Growth characteristics of group A streptococci in a new chemically defined medium. Infect Immun 27, 444–8 (1980).
Takamatsu, D., Osaki, M. & Sekizaki, T. Thermosensitive suicide vectors for gene replacement in Streptococcus suis. Plasmid 46, 140–8 (2001).
Acknowledgements
This study was supported by the National Basic Research Program of China (973 Program grant No. 2012CB518802 for RZ), the International S & T Cooperation Program of China (ISTCP grant No. 2013DFG32360 for RZ), the Biotechnology and Biological Sciences Research Council (BBSRC grant No. BB/L003902/1 for AT) and National Natural Science Foundation of China (31502094 for TZ). We are grateful to Dr. Yosuke Murakami for his pSET plasmids.
Author information
Authors and Affiliations
Contributions
T.Z., J.Z., A.T., H.S. and R.Z. conceived and designed this project and experiments. T.Z., J.Z., S.W. and Q.L. performed the experiments. T.Z., L.L. and S.L. analysed the data and contributed to the development of the figures and tables. T.Z. and R.Z. wrote this manuscript. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhang, T., Zhu, J., Wei, S. et al. The roles of RelA/(p)ppGpp in glucose-starvation induced adaptive response in the zoonotic Streptococcus suis. Sci Rep 6, 27169 (2016). https://doi.org/10.1038/srep27169
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep27169
- Springer Nature Limited
This article is cited by
-
Probiogenomics of Leuconostoc Mesenteroides Strains F-21 and F-22 Isolated from Human Breast Milk Reveal Beneficial Properties
Probiotics and Antimicrobial Proteins (2023)
-
How Streptococcus suis escapes antibiotic treatments
Veterinary Research (2022)
-
Draft genome sequence of Parvularcula flava strain NH6-79 T, revealing its role as a cellulolytic enzymes producer
Archives of Microbiology (2020)