Abstract
The necrotrophic fungal pathogen Sclerotinia trifoliorum exhibits ascospore dimorphism and unidirectional mating type switching - self-fertile strains derived from large ascospores produce both self-fertile (large-spores) and self-sterile (small-spores) offsprings in a 4:4 ratio. The present study, comparing DNA sequences at MAT locus of both self-fertile and self-sterile strains, found four mating type genes (MAT1-1-1, MAT1-1-5, MAT1-2-1 and MAT1-2-4) in the self-fertile strain. However, a 2891-bp region including the entire MAT1-2-1 and MAT1-2-4 genes had been completely deleted from the MAT locus in the self-sterile strain. Meanwhile, two copies of a 146-bp direct repeat motif flanking the deleted region were found in the self-fertile strain, but only one copy of this 146-bp motif (a part of the MAT1-1-1 gene) was present in the self-sterile strain. The two direct repeats were believed to be responsible for the deletion through homologous intra-molecular recombination in meiosis. Tetrad analyses showed that all small ascospore-derived strains lacked the missing DNA between the two direct repeats that was found in all large ascospore-derived strains. In addition, heterokaryons at the MAT locus were observed in field isolates as well as in laboratory derived isolates.
Similar content being viewed by others
Introduction
Fungi have evolved a remarkable diversity of reproductive strategies adapting to the changing environments. Most fungi are able to reproduce both sexually and asexually. Under favorable environmental conditions, fungi may preferentially clonally expand by asexual reproduction. However, when the environmental conditions become adverse some fungi will reproduce sexually to generate genetic variation via recombination and/or resilient surviving structure in the population to enhance and ensure their survival.
In most filamentous ascomycetes, sexual reproduction is mainly controlled by two idiomorphs, MAT1-1 and MAT1-21,2. MAT1-1 encodes a transcription factor with an alpha-1 domain. MAT1-2 encodes a protein with a high mobility group (HMG) DNA-binding domain. The mating-type genes are dissimilar in sequence but found at the same locus on the chromosome and generally flanked by the DNAlyase (APN2) and Cytoskeleton assembly (SLA2). Depending on species, some fungi may contain additional genes in the MAT locus.
Several modes of sexual reproductions in ascomycetes have been reported including homothallic (self-fertile), heterothallic (requires a mating partner, one locus with two alleles), pseudohomothallic (a single ascospore with two nuclei of opposite mating type allele)2,3,4,5, dual mating (isolates can mate with both MAT1-1 and MAT1-2 tester strains) and unidirectional mating type switching (self-fertile strains produce both self-fertile and self-sterile strains)2,3,6,7.
The ascomycetous genus Sclerotinia has three economically important species including Sclerotinia minor, S. sclerotiorum and S. trifoliorum. S. minor and S. sclerotiorum are homothallic, in which single ascospore-derived strains are able to self-fertilize under controlled laboratory conditions and produce ascospores that are fairly uniform in size. However, the ascospores of S. trifoliorum always show size dimorphism with large and small ascospores (4:4 segregation)8,9. Ascospore dimorphism was reported in S. trifoliorum in a taxonomical context as early as in 1954, but was initially considered to be due to heterokaryosis10. It was in 1979 that ascospore size dimorphism was considered to be a character of the species11. Uhm and Fujii8 showed that strains derived from large ascospores are self-fertile (designated L mating type). L mating type strains can produce large and small ascospores with 4:4 segregation8. In contrast, strains derived from small ascospores are self-sterile (designated S mating type). S mating type strains, although self-sterile, can be fertile when spermatized with microconidia from L mating type strains and produce large and small ascospores in 4:4 segregation8,9. This phenomenon of a self-fertile strain giving rise to both self-fertile and self-sterile strains in S. trifoliorum, as well as in other fungal genera, was termed unidirectional mating type switching12. Beside in S. trifoliorum, unidirectional mating type switching occurs in three other fungal genera all in the class Sordariomycetes: Ceratocystis, Chromocrea and Glomerella8,12,13,14,15. Compared to the other sexual reproduction systems in fungi, such as homothallism and heterothallism, the mechanisms of unidirectional mating type switching are less studied. Recent studies have shed some light on the possible mechanisms. Witthuhn et al.15, using PCR specific for the HMG-DNA binding domain, showed that loss of MAT1-2 idomorph was associated with mating type switching in Ceratocystis spp. Wilken et al.14 completely characterized the MAT locus sequences and demonstrated that two 260-bp direct repeats flanking a 3581-bp DNA region caused deletion of this flanked DNA region including MAT1-2 genes in C. fimbriata. They provided the first sequence evidence that direct repeat-mediated deletion results in loss of MAT1-2 genes and in self sterility14.
The structure of the mating type locus in S. minor and S. sclerotiorum has been characterized16,17. Although both S. sclerotiorum and S. minor are homothallic, they have two mating type alleles differentiated by a natural inversion of about 3.6-kb DNA, designated as Inv− and Inv+ MAT alleles16,17. Each allele contains four MAT genes including MAT1-1-1, MAT1-1-5, MAT1-2-1 and MAT1-2-4. Compared to the Inv− MAT allele, about a 3.6-kb region was inverted in the Inv+ MAT allele, which affected orientation of three of the four MAT genes and caused truncation at the 3′ end of MAT1-1-1 and inversion of MAT1-2-4 and MAT1-2-1. However, the MAT locus structure in S. trifoliorum is not known and it was speculated that the mating type switch in S. trifoliorum was related the MAT gene inversion found in S. sclerotiorum and S. minor16,17.
Sclerotinia trifoliorum is not only unique among Sclerotinia spp in producing dimorphic ascospores, but also an important plant pathogens of many cool season legume crops such as alfalfa, clover and chickpea8,11. In order to determine whether the mating type switching in S. trifoliorum is caused by mating type allele inverstion or by DNA deletion, this study was carried out with three objectives: 1) characterize the structure of the MAT locus in the L and S mating types of S. trifoliorum and uncover the possible mechanisms of the unidirectional mating type switching; 2) compare MAT locus with that of S. minor and S. sclerotiorum to better understand the evolution of MAT alleles in Sclerotinia; and 3) develop a screening technique for mating type determination of S. trifoliorum. The results provide further insight into the molecular basis of unidirectional mating type switching and the DNA affected during this evolutionary process.
Results
The structure of the MAT locus in S. trifoliorum self-fertile L mating type most resembles that of MAT inversion positive strains of S. minor and S. sclerotiorum
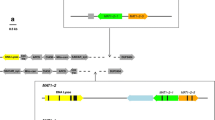
The genome of S. trifoliorum isolate 06CWM-G22 (hereafter G22) was sequenced to 53× coverage using the PacBio platform and the assembled genome of about 39.8 Mb consisted of 20 contigs with N50 length at 2.23 Mb. Telomeres could be detected at both ends of many of the contigs, suggesting the contigs likely represent chromosomes. Contig 3 of about 3.14 Mb was identified to contain the MAT locus (about 16 kb) based on CLC Genomics Workbench searches using the MAT allele (JQ815884) of S. sclerotiorum as query. Five open reading frames (ORF) were predicted within this locus using AUGUSTUS. The first ORF encodes a protein with a high level of similarity to the APN2. The products predicted for the other four complete ORFs showed similarity to MAT1-1-5, MAT1-2-1, MAT1-2-4 and SLA2 and an ORF with high similarity to MAT1-1-1 was divided into two parts by MAT1-2-1 and MAT1-2-4 (Fig. 1). The gene order and orientation in the MAT locus of self-fertile isolate mostly resemble those reported for the Inv+ strains of S. minor and S. sclerotiorum, including truncation of the MAT1-1-1 gene. The only difference is in the translational orientation of the truncated 3′ fragment of the MAT1-1-1; it is in the same orientation as the 5′ fragment of MAT1-1-1 in S. trifoliorum, but in an opposite orientation in S. minor and S. sclerotiorum (Fig. 1)16,17.
Schematic drawing showing the arrangements and orientations of genes in the mating type locus of S. trifoliorum self-fertile L mating type (G22) and self-sterile S mating type (G27) isolates.
The Sclerotinia sclerotiorum Inv+ MAT allele (44Ba12, adapted from Chitrampalam et al.17) is included for comparison. Note the different orientations of the 3′ fragment of MAT1-1-1 gene. The DNA region amplified using the diagnostic primer pair 4F and 6R is indicated which will amplify 3800 bp and 800 bp in the L and S mating types, respectively. Diagram not drawn to scale.
A 2966-bp DNA region was missing in the MAT locus of the self-sterile S mating type
The overlapping 13 pairs of PCR primers designed based on genome sequence were used to amplify DNA fragments from genomic DNA of strains G22 (self-fertile) and 06CWM-G27 (self-sterile, hereafter G27) covering the entire MAT locus including the flanking genes APN2 and SLA2 (see Supplementary Fig. S1). Each pair of the PCR primers was able to amplify expected PCR product from strain G22. Sequences of the PCR products were confirmed by Sanger sequencing. The assembled sequence from the 13 overlapping PCR products from G22 was 16.3 kb long and showed 100% sequence identity to that region in the draft genome, suggesting the quality of the draft genome is highly reliable. However, only 10 of the 13 primer pairs produced expected fragments in the PCR amplifications from strain G27 and the nucleotide sequences of the amplified PCR products were identical to those of strain G22. The remaining three primer pairs (4F/4R, 5F/5R and 6F/6R) did not produce any PCR products from the self-sterile isolate G27. When PCR primers 4F and 6R were paired in PCR, a single PCR product of about 800 bp was amplified, which was subsequently sequenced. This primer pair would produce a PCR product of about 3800 bps in strain G22. The assembled sequence for the G27 MAT locus was 13.9 kb long, 2966-bp shorter than that found in the strain G22 and contained four predicted ORFs, which showed high similarity to APN2, MAT1-1-5, MAT1-1-1 and SLA2 (Fig. 1). The entire MAT1-2-1 and MAT1-2-4 genes were missing along with the deleted 2966-bp DNA region (Fig. 1). The DNA sequences of the MAT locus of self-fertile and self-sterile strains were deposited at GenBank and assigned accession numbers KU726096 and KU726097, respectively.
A 146-bp repeat motif flanked the missing DNA region in the self-fertile L mating type strain
Comparison between the two MAT alleles in the S. trifoliorum strains G22 and G27 revealed a 2966-bp region was missing in the S. trifoliorum strain G27 and the missing DNA region contained the entire MAT1-2-1 and MAT1-2-4 genes. RepFind analysis with a p-value of 0.00 identified a 146-bp direct repeat in the S. trifoliorum G22 MAT locus that appeared twice and located at the boundaries of the deletion region. The first copy of the 146-bp repeat is located in and a part of the MAT1-1-1 gene, whereas the second copy of the repeat is located in a non-coding region. This repeat sequence appeared only once in the S allele strain G27 and is in the MAT1-1-1 gene. The sequence immediately upstream of the repeat motif in the S allele was 100% identical to that upstream of the first copy of the repeat motif in the L allele, whereas the sequence immediately downstream of the repeat motif in the S allele was 100% identical to that downstream of the second copy of the repeat motif in the L allele (Fig. 2). These sequence comparisons strongly suggest that the deletion in the S allele was resulted from intra-molecular homogolous recombination anchored at the two repeats in the L allele, similar to that reported for C. fimbriata14.
Alignment of the three repeat sequences plus 50 bp upstream and 50 bp downstream at the MAT locus Sclerotinia trifoliorum, the first copy (L1) and second copy (L2) of the repeats in the L MAT allele (self-fertile) and the single copy (S) of the repeat in the S MAT allele (self-sterile).
The repeat sequences L1, L2 and S (in red blocks) are 100% identical. The 50 bp upstream of the S repeat is 100% identical to that upstream of the L1 repeat and the 50 bp downstream of the S repeat is 100% identical to that downstrwam of the L2 repeat (identical sequences outside of the repeat sequences are in blue blocks).
Previous studies showed that 250-bp and 256-bp inverted repeat sequences were found in the S. sclerotiorum and S. minor MAT locus, respectively16,17. Alignment of these repeat sequences from the three Sclerotinia species showed the repeat motif in S. trifoliorum was highly similar to part of the other two repeat sequences, although they were different in length (Fig. 3). The repeat motif in S. trifoliorum showed 88.4 and 89.0% identical in nucleotides with the respective repeat motifs in S. minor and S. sclerotiorum, respectively.
Alignment of the direct repeat motif region of self-sterile isolate of Sclerotinia trifoliorum (St, strain G27, 146 bases) with the first copy of the inverted repeat of S. sclerotiorum (Ss, strain SS44Ba1, 250 bases) and S. minor (Sm, strain SM1, 256 bases).
Repeat motifs are in black background and *indicates identical nucleotide among the three species.
Characterization of MAT genes in S. trifoliorum
The MAT1-1-5 gene was 1306-bp long and was identical in both strains G22 and G27 (Table 1). The MAT1-1-1 was 1212-bp long and continuous in the self-sterile strain G27, but was split into two pieces (5′ fragment, 677-bp and 3′ fragment, 601-bp) in the self-fertile strain G22. The MAT1-2-1 and MAT1-2-4 ORFs, present only in the self-fertile strain G22, were 1098 and 879 bp long, respectively. All the MAT genes of S. trifoliorum differed from their respective MAT genes in S. minor strain SM116, S. sclerotiorum strains 198018 and 44Ba117 and B. cinerea strain T418. Comparison between the MAT genes of S. trifoliorum to their corresponding MAT genes in S. minor, S. sclerotiorum and B. cinerea indicated 43% to 95% nucleotide identity depending on the MAT gene (Table 1). MAT1-1-1 was 90, 89 and 77% identical to the corresponding genes in S. minor, S. sclerotiorum and B. cinerea, respectively. MAT1-1-5 was 95, 95 and 77% identical to the corresponding genes in S. minor, S. sclerotiorum and B. cinerea, respectively. MAT1-2-1 was 94, 94 and 78% identical to the genes in S. minor, S. sclerotiorum and B. cinerea, respectively. Among the four MAT genes, MAT1-2-4 was the most divergent with the lowest sequence identity (43%) to B. cinerea homologs. MAT1-1-5, MAT1-2-1 and MAT1-2-4 in isolate G22 contains 3, 2 and 1 introns, respectively and MAT1-1-5 and MAT1-1-1 in isolate G27 contains 3 and 2 introns, respectively (Table 1). Sequencing RT-PCR products showed that the intron sequences were absent in the RT-PCR products.
Analysis of the intergenic spacer regions between MAT genes in Sclerotinia spp
The arrangement of MAT genes and their translation directions in the L mating type strain of S. trifoliorum were most similar to those of the Inv+ MAT allele of S. minor and S. trifoliorum. Therefore, the intergenic regions of S. trifoliorum MAT locus were compared with those in the Inv+ MAT allele of S. minor and S. trifoliorum. Except for the spacer region between MAT1-2-1 and MAT1-2-4, significant difference in length in all other MAT intergenic regions were found among the three Sclerotinia spp. (Table 2). Similar to S. sclerotiorum and S. minor, the intergenic region between APN2 and MAT1-1-5 (2917 bp) is the longest among the noncoding intergenic regions at the MAT locus in S. trifoliorum and it is 20 and 1318 bp longer than its corresponding regions in S. sclerotiorum and S. minor, respectively. The intergenic region between MAT1-1-5 and the 5′ fragment of MAT1-1-1 is 2851 bp, which is 2445 bp longer than its homologue in S. sclerotiorum and S. minor. A 12-bp intergenic region between the 5′ fragment of MAT1-1-1 and MAT1-2-1 in strain G22 were identified, but no such a spacer region was found in the Inv+ MAT allele of S. sclerotiorum and S. minor. The intergenic region between MAT1-2-4 and 3′ fragment of MAT1-1-1 is 177-bp long and 330-bp shorter than its homologue in S. sclerotiorum and S. minor. The intergenic region between the 3′ fragment of MAT1-1-1 and SLA2 is 1292-bp long and 676 and 668 bp longer than its homologue in S. sclerotiorum and S. minor, respectively.
Comparison of MAT proteins
The analysis of amino acid sequences indicated that MAT proteins in S. trifoliorum were 73–92% identical to the respective proteins in S. minor, 64–91% identical to those of the respective proteins in S. sclerotiorum and 51-77% identical to the corresponding proteins in B. cinerea. Among the four MAT proteins of S. trifoliorum, MAT1-2-4 protein exhibited the most divergent with lowest identity (51–74%) to the homologs in S. minor, S. sclerotiorum and B. cinerea.
Comparison of the conserved alpha-box (for MAT1-1-1) and HMG-box (for MAT1-2-1) of S. trifoliorum to the respective domains in S. minor, S. sclerotiorum and B. cinerea revealed that the HMG-box in S. trifoliorum was more similar than the alpha-box to the corresponding domains in other species (Supplementary Fig. S2).
Transcription analysis of MAT genes in S. trifolorium
RT-PCR was carried out to detect expression of the MAT genes in S. trifoliorum strains G22 and G27. Three of the five primer pairs used in the RT-PCR each spanned an intron (Supplementary Table S3), facilitated detection of potential DNA contamination in the RNA preparations. Sequencing of the RT-PCR products did not detect the intron sequences suggesting that there was no DNA contamination. The results showed all four MAT genes (MAT1-1-1, MAT1-1-5, MAT1-2-1 and MAT1-2-4) in S. trifoliorum strain G22 were expressed (Fig. 4). Even the two split fragments of MAT1-1-1 gene were expressed although the 3′ fragment of MAT1-1-1 lacked an in-frame start codon. Expression of MAT1-1-5 and MAT1-1-1 were also detected in the self-sterile strain G27 of S. trifoliorum. However, no transcripts of MAT1-2-1 and MAT1-2-4 genes were detected in the self sterile strain G27 (Fig. 4).
Expression of MAT genes of self-fertile isolate G22 and self-sterile isolate G27 of Sclerotinia trifoliorum.
Primer sequences and expected product sizes are shown in Supplementary Table S3 and sequencing of the RT-PCR products showed no intron sequences, suggesting the products were from RNA only. Lane M is DNA ladder 100-bp Hyperladder.
MAT allele determination by PCR and association of the MAT alleles with ascospore size
The two primers (4F and 6R) that flank the deletion region were used to detect and differentiate the L and S MAT alleles in single ascospore isolates and field isolates. Ascospores in two asci (each from a different field strain) were determined to be either large or small spores microscopically at time of ascus dissection. The large and small ascospores were in 4:4 arrangement in one ascus from strain G22 and in 2:2:2:2 arrangement in the other ascus from strain G34. Without exception, PCR reactions with primers 4F and 6R always produced the small amplicon of 800 bps (S MAT allele) in the strains derived from the small ascospores and the large amplicon of 3800 bp (L MAT allele) in the strains derived from large ascospores (Fig. 5AB). Occasionally isolates with L MAT allele also showed a faint PCR product of the S allele, showing potential heterokaryon at the MAT locus. The nature of the faint band as to its origin remains to be investigated.
PCR detection of the MAT alleles of Sclerotinia trifoliorum isolates using the specific primers 4F and 6R.
(A) Mating type alleles of single-ascospore strains from an ascus of isolate 06CWM-G22 with 4 large and 4 small ascospores distriution. Lanes 1, 2, 3 and 4 are strains derived from large ascospores and lanes 5, 6, 7 and 8 are strains derived from small ascospores. (B) Mating type alleles of single ascospore isolates from the an ascus of isolate 06CWM-G34. Lanes 1, 2, 5 and 6 are strains derived from large ascospores and lanes 3, 4, 7 and 8 are strains derived from small sacospores. (C) Mating type alleles of S. trifioliorum field isolates. Lanes in 1-13 are 06CWM-G22, 06CWM-G48, 06CWM-G2, 06CWM-G55, 06CWM-F9, 06CWM-F6, 06CWM-F2, 05WM21, 06CWM-G7, 06CWM-G23, 06CWM-D5, 06CWM-G39 and 06CWM-G27, respectively.
Additionally, the 4F and 6R PCR primers were applied to 22 field isolates to test their applicability to differentiate the MAT alleles in isolates of different genetic backgrounds. Nine of the 22 isolates produced the 0.8 kb product and are S mating type. Thirteen isolates produced the 3.8 kb band, suggesting they are L mating type isolates (Fig. 5C; see Supplementary Table S1). And five of these 13 solates also showed a faint 0.8 kb band, which mean these isolates are likely heterokaryotic with respect to the MAT locus (Fig. 5, see Supplementary Table S1). The heterokaryons are self-fertile and have the phenotype of the L mating type. The field isolates could be unambiguously differentiated into either the L or the S mating types using PCR with primers 4F and 6R.
Discussion
The MAT loci in two homothallic Sclerotinia species S. minor and S. sclerotiorum, close relatives of S. trifoliorum, were recently characterized16,17. Two mating type (MAT) alleles, inversion negative (Inv−) and inversion positive (Inv+) have been reported in these two species. They have typical features of MAT locus in homothallic Pezizomycotinia, in which two idomorphs, MAT1-1 with the alpha domain and MAT1-2 with HMG-domain, are present in tandem in a single MAT locus that is flanked by APN2 and SLA2. In addition, the MAT loci in these two species carry two newly discovered MAT genes, MAT1-1-5 and MAT1-2-4, which are homologues of MAT1-1 and MAT1-2, respectively18. The present study showed the organization of MAT allele in S. trifoliorum G22 was identical to that in the Inv+ MAT allele of both S. sclerotiorum and S. minor, except for the orientation of the truncated 3′ fragment of MAT1-1-1. In S. trifoliorum strain G22, the orientation of the 3′ fragment of MAT1-1-1 is in the same direction as the 5′ fragment of MAT1-1-1. In contrast, the 3′ fragment and the 5′ fragment of the MAT1-1-1 gene were in opposite orientation in the Inv+ MAT allele of both S. sclerotiorum and S. minor16,17. Depending on the MAT genes, 89–95% nucleotide identity was observed among the three Sclerotinia spp. Another similarity in the MAT locus among the Sclerotinia spp. was the presence of repeat motifs of the same origin with significant sequence identities (Fig. 3) and the repeat motif is part of the MAT1-1-1 gene sequence in all three Sclerotinia spp.
Despite these similarities, there are significant differences in repeat motif orientation, in gene sequences and in the noncoding spacer regions at the MAT locus among the Sclerotinia spp, although the functions of noncoding spacer regions during the evolution of mating locus are unknown. The major difference in the MAT locus among the Sclerotinia spp. is the orientation of the repeat motifs. In S. minor and S. sclerotiorum, the repeat motifs are inverted (in opposite direction), whereas in S. trifoliorum, the repeat motifs are direct repeat (in the same direction). During meiosis via recombination, inverted repeats would cause inversion of the DNA region between the repeats16,17, whereas direct repeats would cause deletion of the DNA region between the repeats14. The situation in S. trifoliorum is very similar to that recently reported for the mating type switching mechanism in Ceratocystis fimbriata by Wilken et al.14. Wilken et al. showed that two 260-bp direct repeats flanking the deleted DNA region in self-fertile isolates of C. fimbriata and the DNA region between the two repeats along with one copy of the repeat motif was missing in the self-sterile isolates. Thus the unidirectional mating type switching in S. trifoliorum is caused by the same molecular mechanisms as reported in C. fimbriata: direct repeat-mediated DNA deletion causing loss of MAT-1-2 genes results in self-sterility.
Despite of the same molecular mechanism for the unidirectional mating type switching in both C. fimbriata and S. trifoliorum, this unidirectional mating type switching phenotype has obviously evolved independently. First, the repeat sequences are completely different and there are no sequence similarities between the repeat motifs of C. fimbriata and S. trifoliorum. Second, the locations of the repeat sequences are totally different. The repeat motif is located in and a part of the MAT1-1-1 gene in S. trifoliorum, whereas they are located in non-coding regions in C. fimbriata, Finally, the genes, gene orientations and flanking genes are different. The MAT locus of S. trifoliorum possesses MAT1-1-5 and MAT1-2-4, unique for Sclerotinia and Botrytis, whereas the MAT locus of C. fimbriata possesses MAT1-1-2. The MAT locus in S. trifoliorum is flanked by APN2 and SLA2 genes, typically found in many Ascomycetes, whereas the MAT locus in C. fimbriata is downstream of APN2 and SLA2 genes14.
Homothallism and heterothallism are the two most common strategies of sexual reproduction in fungi. Different hypotheses exist as to which came first1,2. In Sclerotinacae, an early hypothesis suggested that heterothallic Botrytis cinerea evolved from homothallic S. sclerotiorum through four step mutations18. Chitrampalam et al.17 proposed a more parsimonious three-step mutation for evolution of homothallic Sclerotinia spp. from heterothallic Botrytis. Understanding the MAT locus structures in S. trifoliorum helped shed more light on this. The structure of L allele of S. trifoliorum MAT locus most resembles the structure of the Inv+ MAT allele in S. minor and S. sclerotiorum. However, the structure of S allele of S. trifoliorum is the same as in the MAT1-1 allele (idiomorph) in Botrytis cinerea. Imagining an ancestral heterothallic species with MAT locus similar to B. cinerea, a one-step mutation, a crossover would result in insertion of the MAT1-2 allele (idiomorph) into MAT1-1 idiomorph and truncate MAT1-1-1 gene, forming a structure as the L allele of S. trifoliorum (Fig. 6) and the Inv+ MAT allele of S. minor and S. sclerotiorum. Our data support the hypothesis that homothallic Sclerotinia spp evolved from an ancestral heterothallic species.
Just like in S. sclerotiorum and S. minor, the MAT genes were constitutively expressed during vegetative growth. Thus the MAT genes may have other functions besides sexual reproduction. In S. trifoliorum, there is a clear pleiotrophic effect on ascospore size. Lack of the MAT1-2 genes resulted in ascospores of a smaller size. Thus the ascospore size is correlated to the genotype in the spore itself, not the genotype of the parent strain. We compared 20 isolates each derived from large ascospores and small ascospores for mycelial growth and sclerotial production on PDA, but did not observe difference between the two types (unpublished data). However, a study in Ceratocystis albifundus showed that self-sterile isolates (without MAT1-2 idiomorph) have lower fitness in germination, growth rate and pathogenicity than self-fertile isolates19. An earlier study suggested that the two spore types of S. trifoliorum may differ in initial germination rate8. Whether the MAT genes have any other pleiotrophic effect in S. trifoliorum requires further investigation. Although the MAT1-1-1 gene in the L allele was truncated and divided by the MAT1-2 genes and its 3′ fragment lacked an in-frame start codon, both the two divided fragments were successfully transcribed like the other three intact MAT genes (Fig. 4). Similar results were reported in S. minor and S. sclerotiorum, in which the 3′ fragment of the truncated MAT1-1-1 gene was also expressed. Whether the MAT1-1-1 gene could still be functional without expressing its 3′ fragment downstream of the alpha box in a homothallic strain is still unknown.
In this study we also observed heterokayons at the MAT locus (presence of both L and S alleles) in field isolates as well as in laboratory-derived single ascospore isolates (Fig. 5). Interestingly, in all cases, it was the L allele showing the stronger band than the S allele. This unequal PCR product intensity was unlikely due to preferential amplification because the weaker band was always the S allele that is smaller in size than the L allele and smaller sized PCR product would be preferentially amplified. Heterokaryon at MAT locus has also been reported in many filamentous fungi including S. minor and S. sclerotiorum, Botryotinia fuckeliana and S. homoeocarpa16,17,20,21,22,23,24. Especially, the heterokaryosis during the vegetative phase was commonly found in the most isolates of Cryphonectria parasitica collected from the field22,23. Formation of heterokaryons in S. sclerotiorum under nonrestrictive conditions was demonstrated before25 and more than 50% of field isolates of S. minor and S. sclerotiorum were found to be heterokaryotic at the MAT locus16,24. However, heterokaryons were not observed in microsatellite loci among many isolates of S. sclerotiorum26,27. Future study should be directed at determining whether such repeat sequence-mediated DNA deletion or DNA inversion through intramolecular recombination observed during meosis could also occur during mitosis at low frequency, which could have significant implications in interpreting population genetic data.
Material and Methods
Strains and nucleic acid extraction
S. trifoliorum isolates were collected from stems of infected chickpea plants from various locations in central California, USA (see Supplementary Table S1)28. The isolates were maintained at 20 °C on PDA. Long-term storage stock was maintained at −20 °C as sclerotia. Mycelia of 3 to 4 day old culture were harvested from colonies grown on cellophane-covered PDA plate and used for nucleic acid extraction immediately or stored at −20 °C until use. Genomic DNA was extracted using the Qiagen Plant DNA extraction kit (Qiagen) and quantified using a ND-1000 Nanodrop spectrometer (NanoDorp Technologies). The working concentration of DNA for PCR amplification was adjusted to10 ng/μL with sterilized distilled water. Total RNA was extracted from the mycelium of isolates G22 and G27 using the Qiagen RNeasyTM Plant Mini Kit following the manufacturer’s protocol and stored at −20 °C until use.
Tetrad analyses
Apothecia were produced from sclerotia of L mating type strains 06CWM-G22 and 06CWM-G34 following the protocol described by Njambere et al.28. Asci in an apothecium were collected by crashing the apothecia and diluting in 1 mL water, then spread on one end of a microscope slide coated with 3% water agar8. Asci were dissected with a micromanipulator and individual ascospores were isolated from the apex of the ascus. Ascospores were transferred to the opposite end of the agar-coated slide and placed at a distance apart from each other in the order of their original arrangement in the ascus and allowed to germinate. After 12 h, the germinated spores were transferred to PDA plates under a dissecting microscope and maintained as individual strains.
Genome sequencing and MAT locus re-sequencing
The genome of the L mating type strain 06CWM-G22 was sequenced using the PacBio RS II on eight SMRT cells at University of Washington and assembled at the Washington State University. After identifying the contig where the MAT locus was located (see In silico analyses below), 13 primers pairs were designed based on the draft genome sequence to amplify and cover the entire locus including the flanking genes APN2 and SLA2 (see Supplementary Fig. S1 and Supplementary Table S2). These 13 primer pairs were used in PCR amplification of strain G22 of L mating type and strain G27 of S mating type. The following PCR cycling conditions were used for each of the 13 primer pairs: initial denaturation (94 °C for 4 min); 35 cycles of denaturation (94 °C for 30 s), annealing (55 °C for 30 s) and extension (68 °C, 30 s); final extension (68° for 8 min); hold (15 °C). Reaction mixtures were 20 μL in volume and contained 1.0 × of 10 × PCR buffer, 1.5 mM MgSO4, 0.5 mM total dNTPs, 0.5 μM primer (each), 0.5 U Taq and 2 μL of 10 ng/μL template DNA. The PCR products were checked using 1% agarose gel electrophoresis (IBC BioExpress), purified with ExoSAP-IT and sequenced using corresponding primers at the Washington State University Bioinformatics Core, Pullman, WA, USA. The overlapping sequences from the PCR amplicons were assembled and analyzed in CLC Genomics Workbench.
In silico analyses
To identify the contig in the draft genome that contained the MAT locus, the MAT locus (JQ815884) of S. sclerotiorum was used as query to search the draft genome sequence of isolate G22 using the CLC Genomics Workbench. The MAT coding and intron positions of S. trifoliorum were predicted using the de novo AUGUSTUS online interface. For AUGUSTUS, predictions were derived from the available model that was trained on Botrytis cinerea. Protein domains were identified with searches of the full pfam database V27.0 (http://pfam.xfam.org/). To detect the repeat sequence in the MAT locus sequences of G22 and G27, Tandem Repeat Finder was used to examine the MAT locus sequences against themselves29.
Comparison of the MAT locus and gene arrangements
To determine the L and S alleles of the MAT locus in multiple isolates, a pair of primers (4F/6R) was selected from the 13 primer pairs to flank the sequence of the deletion region. All isolates used in this study were subjected to PCR amplification with the primers. The PCR conditions are same as described above. The PCR product sizes in L and S alleles were about 3.8 kb and 0.8 kb, respectively. The PCR fragments were checked by agarose gel electrophoresis and PCR products of representative isolates were sequenced as described above to confirm sequence identity.
Gene Expression analysis
RT-PCR assays were used to monitor the expression of MAT genes in strain G22 of L mating type and strain G27 of S mating type. First-strand cDNA synthesis was prepared from DNase-treated total RNA using the iScript cDNA Synthesis Kit (Bio-Rad) following the manufacturer’s instructions. The house keeping gene β-tubulin was included as a positive control. Five primer pairs for four MAT genes (MAT1-1-5, 5′ truncated MAT1-1-1, MAT1-2-1, MAT1-2-4 and 3′ truncated MAT1-1-1) were listed in Supplementary Table S3. The RT-PCR primer pairs for transcripts of MAT1-1-5, 5′ fragment of MAT1-1-1 and MAT1-2-1 each span an intron, to allow detection of potential DNA contamination and to confirm correct intron splicing by sequencing. The RT-PCR products for each gene was checked by electrophoresis and also by sequencing using the corresponding PCR primers to confirm the gene-specific amplification and appropriate intron locations.
Additional Information
How to cite this article: Xu, L. et al. Direct repeat-mediated DNA deletion of the mating type MAT1-2 genes results in unidirectional mating type switching in Sclerotinia trifoliorum. Sci. Rep. 6, 27083; doi: 10.1038/srep27083 (2016).
References
Yun, S. H., Berbee, M. L., Yoder, O. C. & Turgeon, B. G. Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proceedings of the National Academy of Sciences of the United States of America 96, 5592–5597 (1999).
Ni, M., Feretzaki, M., Sun, S., Wang, X. & Heitman, J. Sex in fungi. Annual review of genetics 45, 405–430, 10.1146/annurev-genet-110410-132536 (2011).
Coppin, E., Debuchy, R., Arnaise, S. & Picard, M. Mating types and sexual development in filamentous ascomycetes. Microbiology and molecular biology reviews : MMBR 61, 411–428 (1997).
Nelson, M. A. Mating systems in ascomycetes: a romp in the sac. Trends in genetics: TIG 12, 69–74 (1996).
Raju, N. B. & Perkins, D. D. Programmed ascospore death in the homothallic ascomycete Coniochaeta tetraspora. Fungal genetics and biology : FG & B 30, 213–221, 10.1006/fgbi.2000.1217 (2000).
Putman, A. I., Tredway, L. P. & Carbone, I. Characterization and distribution of mating-type genes of the turfgrass pathogen Sclerotinia homoeocarpa on a global scale. Fungal genetics and biology: FG & B 81, 25–40, 10.1016/j.fgb.2015.05.012 (2015).
Haber, J. E. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191, 33–64, 10.1534/genetics.111.134577 (2012).
Uhm, J. Y. & Fujii, J. Ascospore dimorphism in Sclerotinia trifoliorum and cultural characters of strains from different-sized spores. Phytopathology 73, 565–569 (1983).
Uhm, J. Y. & Fujii, J. Heterothallism and mating type mutation in Sclerotinia trifoliorum. Phytopathology 73, 569–572 (1983).
Carr, A. J. H. In Proceedings of the 8th International Botanical Congress Vol. Section 19 72–74 (Paris, France., 1954).
Kohn, L. M. Delimitation of the economically important pathogenic Sclerotinia species. Phytopathology 69, 881–886 (1979).
Perkins, D. D. Mating-type switching in filamentous ascomycetes. Genetics 115, 215–216 (1987).
Harrington, T. C. & McNew, D. L. Self-fertility and uni-directional mating-type switching in Ceratocystis coerulescens, a filamentous ascomycete. Current genetics 32, 52–59 (1997).
Wilken, P. M., Steenkamp, E. T., Wingfield, M. J., de Beer, Z. W. & Wingfield, B. D. DNA loss at the Ceratocystis fimbriata mating locus results in self- sterility. PloS one 9, 10.1371/journal.pone.0092180 (2014).
Witthuhn, R. C., Harrington, T. C., Wingfield, B. D., Steimel, J. P. & Wingfield, M. J. Deletion of the MAT-2 mating-type gene during uni-directional mating-type switching in Ceratocystis. Current genetics 38, 48–52 (2000).
Chitrampalam, P. & Pryor, B. M. Characterization of mating type (MAT) alleles differentiated by a natural inversion in Sclerotinia minor. Plant Pathol 64, 911–920, 10.1111/ppa.12305 (2015).
Chitrampalam, P., Inderbitzin, P., Maruthachalam, K., Wu, B. M. & Subbarao, K. V. The Sclerotinia sclerotiorum mating type locus (MAT) contains a 3.6-kb region that is inverted in every meiotic generation. PloS one 8, e56895, 10.1371/journal.pone.0056895 (2013).
Amselem, J. et al. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet 7, e1002230, 10.1371/journal.pgen.1002230 (2011).
Lee, D. H., Roux, J., Wingfield, B. D. & Wingfield, M. J. Variation in growth rates and aggressiveness of naturally occurring self-fertile and self-sterile isolates of the wilt pathogen Ceratocystis albifundus. Plant Pathol 64, 1103–1109, 10.1111/ppa.12349 (2015).
Liberti, D., Rollins, J. A. & Harmon, P. F. Evidence for morphological, vegetative, genetic and mating-type diversity in Sclerotinia homoeocarpa. Phytopathology 102, 506–518, 10.1094/Phyto-06-11-0180 (2012).
Faretra, F. & Pollastro, S. Genetic studies of the phytopathogenic fungus Botryotinia fuckeliana (Botrytis cinerea) by analysis of ordered tetrads. Mycol Res 100, 620–624 (1996).
McGuire, I. C. et al. Heterokaryon formation and parasexual recombination between vegetatively incompatible lineages in a population of the chestnut blight fungus, Cryphonectria parasitica. Molecular ecology 14, 3657–3669, 10.1111/j.1365-294X.2005.02693.x (2005).
McGuire, I. C., Marra, R. E. & Milgroom, M. G. Mating-type heterokaryosis and selfing in Cryphonectria parasitica. Fungal Genet Biol 41, 521–533, 10.1016/j.fgb.2003.12.007 (2004).
Chitrampalam, P. et al. Prevalence of inversion positive and inversion negative mating type (MAT) alleles and MAT heterokaryons in Sclerotinia sclerotiorum in the United States. Botany 93, 497–505, 10.1139/cjb-2015-0035 (2015).
Ford, E. J., Miller, R. V., Gray, H. & Sherwood, J. E. Heterokaryon formation and vegetative compatibility in Sclerotinia sclerotiorum. Mycol Res 99, 241–247 (1995).
Attanayake, R. N., Carter, P. A., Jiang, D., Del Rio-Mendoza, L. & Chen, W. Sclerotinia sclerotiorum populations infecting canola from China and the United States are genetically and phenotypically distinct. Phytopathology 103, 750–761, 10.1094/PHYTO-07-12-0159-R (2013).
Attanayake, R. N. et al. Inferring outcrossing in the homothallic fungus Sclerotinia sclerotiorum using linkage disequilibrium decay. Heredity 113, 353–363, 10.1038/hdy.2014.37 (2014).
Njambere, E. N. et al. Stem and crown rot of chickpea in California caused by Sclerotinia trifoliorum. Plant Dis 92, 917–922, 10.1094/Pdis-92-6-0917 (2008).
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic acids research 27, 573–580 (1999).
Acknowledgements
We thank two anonymous reviewers for critically evaluate an earlier version of the manuscript and helpful suggestions and Dr. Daniel Mullendore from the Franceschi Microscopy and Imaging Center at Washington State University for technical support. The research was funded in part by the USDA ARS National Sclerotinia Initiative.
Author information
Authors and Affiliations
Contributions
L.X. and W.C. designed the experiments. L.X. and T.J. performed the experiments. L.X. and W.C. analyzed the data and wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Xu, L., Jardini, T. & Chen, W. Direct repeat-mediated DNA deletion of the mating type MAT1-2 genes results in unidirectional mating type switching in Sclerotinia trifoliorum. Sci Rep 6, 27083 (2016). https://doi.org/10.1038/srep27083
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep27083
- Springer Nature Limited
This article is cited by
-
Sclerotinia sclerotiorum populations: clonal or recombining?
Tropical Plant Pathology (2019)
-
Phenotypic and genotypic characterization of single isolate-derived monoascospore strains of Sclerotinia sclerotiorum from common bean
Tropical Plant Pathology (2019)
-
Characterization of mating-type idiomorphs suggests that Morchella importuna, Mel-20 and M. sextelata are heterothallic
Mycological Progress (2017)