Abstract
Recently, we reported the isolation of Streptobacillus hongkongensis sp. nov. from patients with quinsy or septic arthritis. In this study, we developed a PCR sequencing test after sulfamethoxazole/trimethoprim and nalidixic acid enrichment for detection of S. hongkongensis. During a three-month study period, among the throat swabs from 132 patients with acute pharyngitis and 264 controls, PCR and DNA sequencing confirmed that S. hongkongensis and S. hongkongensis-like bacteria were detected in 16 patients and 29 control samples, respectively. Among these 45 positive samples, five different sequence variants were detected. Phylogenetic analysis based on the 16S rRNA gene showed that sequence variant 1 was clustered with S. hongkongensis HKU33T/HKU34 with high bootstrap support; while the other four sequence variants formed another distinct cluster. When compared with the 16S rRNA gene of S. hongkongensis HKU33T, the five sequence variants possessed 97.5–100% sequence identities. Among sequence variants 2–5, their sequences showed ≥99.5% nucleotide identities to each other. Forty-two individuals (93.3%) only harbored one sequence variant. We showed that the human oropharynx is a reservoir of S. hongkongensis, but the bacterium is not associated with acute pharyngitis. Another undescribed novel Streptobacillus species is probably also residing in the human oropharynx.
Similar content being viewed by others
Introduction
Streptobacillus is one of the four genera within the family Leptotrichiaceae. It was first isolated from the blood of a patient who suffered from a rat bite in 19141. Since the establishment of the genus Streptobacillus in 1925, this genus had contained only one single species, Streptobacillus moniliformis, for almost 90 years2. S. moniliformis is the causative agent of streptobacillary rat-bite fever3. It is also associated with amnionitis4, bacteremia5,6,7, brain abscess8, cutaneous and subcutaneous abscesses9,10, endocarditis11,12, female genital tract abscess13, palmoplantar pustulosis14, septic arthritis15,16,17, spinal epidural abscess18, splenic abscess19, spondylodiscitis with psoas abscess20, and synovitis21. Microbiologically, S. moniliformis is a Gram-negative facultative anaerobic bacillus that grows in chains. It is naturally harbored in the oral cavity22 and upper respiratory tract23,24 of rats.
In 2014, we reported the isolation of a novel Streptobacillus species, named S. hongkongensis, from the pus of a patient with quinsy and the elbow joint fluid of another patient with tophaceous gout and left elbow septic arthritis25. Both strains of S. hongkongensis were resistant to cotrimoxazole and nalidixic acid as determined by disk diffusion test (unpublished data). Since S. moniliformis colonizes the oral cavity of rats while S. hongkongensis could be isolated from peritonsillar abscess pus of human, we hypothesize that the oropharynx of human may be the natural reservoir of this bacterium or S. hongkongensis may be associated with acute pharyngitis. To test these hypotheses, we first developed an in-house molecular test for the detection of S. hongkongensis on throat swabs of patients with acute pharyngitis. Preliminary study showed that S. hongkongensis or S. hongkongensis-like sequences were detected in around 10% of 100 patient samples collected (unpublished data), indicating the presence of this bacterium in human oropharynx. In this molecular epidemiological study, in order to further test for the association of S. hongkongensis with acute pharyngitis, we used the in-house developed molecular method to examine for any significant difference between the detection rates of S. hongkongensis on throat swabs of patients with acute pharyngitis and healthy controls.
Results
Antimicrobial susceptibility
S. hongkongensis HKU33T and HKU34 grew as white clumps of cells in brain-heart infusion (BHI) broth after 3 days of incubation under aerobic condition supplemented with 5% CO2 at 37 °C. Growth was observed for both strains at all drug concentrations tested for both sulfamethoxazole/trimethoprim and nalidixic acid. Sulfamethoxazole/trimethoprim at a concentration of 400/80 μg/ml and nalidixic acid at a concentration of 40 μg/ml were used for enrichment purpose.
Primer specificity
PCR of the partial 16S rRNA gene of S. hongkongensis strains HKU33T and HKU34 using the primer pair LPW21593/LPW21594 yielded DNA fragments of about 700 bp with strong signal. For S. moniliformis CCUG 13453T, Sneathia sanguinegens CCUG 41628T, and “Sneathia amnii” CCUG 52976, no PCR product of expected size was detected (Fig. 1).
Molecular detection of S. hongkongensis and S. hongkongensis-like bacteria
During the three-month period, 132 throat swab samples from 132 patients [male:female = 7:4, age (median, range) = 18.5 years, 0–92 years] with acute pharyngitis were sent to our clinical microbiology laboratory and 264 throat swab samples from 264 controls [male:female = 85:47, age (median, range) = 23 years, 0–95 years], including 132 healthy individuals and 132 out-patients not on antibiotics, without acute pharyngitis were included. Among the 132 patient and 264 control samples collected, PCR and DNA sequencing confirmed that S. hongkongensis and S. hongkongensis-like bacteria were detected in 16 patient and 29 control samples (Table 1).
Sequence analysis and phylogenetic characterization
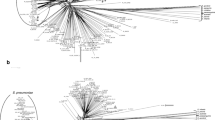
Among all of the 45 positive samples, five different sequence variants were detected. Phylogenetic analysis based on the 16S rRNA gene showed that sequence variant 1 was clustered with S. hongkongensis HKU33T and HKU34 with high bootstrap support; while the other four sequence variants formed another distinct cluster (Fig. 2). When compared with the 16S rRNA gene of S. hongkongensis HKU33T, sequence variants 1, 2, 3, 4, and 5 possessed 100%, 97.5%, 97.7%, 97.5%, and 97.7% sequence identities, respectively. Among sequence variants 2, 3, 4, and 5, their sequences showed ≥99.5% nucleotide identities. Forty-two individuals (93.3%) only harbored one sequence variant (variants 1, 2 and 4). However, in one patient and one control sample, two sequence variants (variants 2 and 4) were detected while in another control sample all the five sequence variants were detected (variants 1–5).
The tree was inferred from 16S rRNA gene sequence data by the maximum likelihood method with the substitution model K2 (Kimura 2-parameter model) + G (gamma-distributed rate variation) + I (estimated proportion of invariable sites), and was rooted using Fusobacterium mortiferum DSM 19809T. The scale bar indicates the estimated number of substitutions per base. Numbers at nodes, expressed in percentage, indicate levels of bootstrap support calculated from 1,000 trees and bootstrap values lower than 70 are not shown. All accession numbers (in parentheses) are given as cited in the DDBJ/ENA/GenBank databases. The five S. hongkongensis sequence variants reported in this study are highlighted in bold type.
Discussion
In this study, we showed that human oropharynx is a reservoir of S. hongkongensis. Since numerous bacterial species reside in the human oropharynx, we developed a PCR test after sulfamethoxazole/trimethoprim and nalidixic acid enrichment, as the two S. hongkongensis strains we isolated previously, HKU33T and HKU34, were resistant to these antibiotics. These antibiotics were also widely used as enrichment supplements for the detection of Streptobacillus-like bacteria previously22,26. Using this molecular method for detection, overall, S. hongkongensis (sequence variant 1), which possessed a 100% 16S rRNA gene sequence identity with those of the two previously isolated S. hongkongensis strains HKU33T and HKU3425, was found in 2.0% of the throat swabs from all subjects tested. There was no significant difference between the detection rates for patients and controls, indicating that the bacterium is not associated with acute pharyngitis. All positive samples (sequence variant 1) were collected from children under the age of 14 years (p < 0.002 by Fisher’s exact test) (Table 1) without any sex predilection, suggesting children are the major population carrying the bacterium. On the other hand, it is notable that the two strains of S. hongkongensis which we previously isolated were recovered from the peritonsillar abscess pus of a 38-year-old man and the joint fluid of a 64-year-old man, respectively25, indicating that diseases caused by S. hongkongensis can occur in adults. Isolation and identification of more strains of S. hongkongensis from infective sites would reveal a more detail epidemiology and disease spectrum of this bacterium.
Another undescribed novel Streptobacillus species is likely also residing in the human oropharynx. In the present study, in addition to S. hongkongensis (sequence variant 1), four other 16S rRNA gene sequence variants (sequence variants 2–5), which possessed ≥99.5% nucleotide identities among themselves, 97.5–97.7% nucleotide identities to that of S. hongkongensis, and ≤95.9% nucleotide identities to that of the type strains of other Streptobacillus species, were detected. Since these four sequence variants form a unique cluster distinct from S. hongkongensis (sequence variant 1) as well as other known Streptobacillus species, it is likely that this represent another novel Streptobacillus species (Fig. 2). It is notable that the 16S rRNA gene sequences of S. felis and S. notomylis, the two recently described Streptobacillus species, possessed 99.1% nucleotide identity when compared with each other and ≥97.6% nucleotide identities to that of S. moniliformis27,28. However, subsequent DNA-DNA hybridization experiment and pairwise whole genome comparison showed that they are two separate species distinct from S. moniliformis27,28. Since the potentially novel Streptobacillus species (sequence variants 2–5) can be detected by the present molecular test, it should also be resistant to sulfamethoxazole/trimethoprim and nalidixic acid. Overall, this potentially novel Streptobacillus species was detected in 9.6% of the throat swabs from all subjects tested, suggesting that its prevalence in human oropharynx is higher than that of S. hongkongensis. Similar to S. hongkongensis, this potentially novel Streptobacillus species was also found primarily in subjects under the age of 14 (p < 0.001 by Fisher’s exact test), particularly in children below five years of age (p < 0.05 by Fisher’s exact test) (Table 1) with no sex predilection.
More novel Streptobacillus species could be discovered from other animals. The genus Streptobacillus had remained monotypic for almost a century2. In the last two decades, the use of 16S rRNA gene sequencing has led to the discovery of numerous novel bacteria29. In the past two years, four novel Streptobacillus species were described, including S. hongkongensis, S. felis, S. notomylis, and S. ratti. S. felis was isolated from the lung of a cat with acute suppurative to fibrinous, focally necrotizing bronchopneumonia with multifocal desquamation of type II pneumocytes and alveolar macrophages28; whereas S. notomylis from the heart of an Australian spinifex hopping mouse (Notomys alexis) with septicemia as well as the oral swabs of rats (Rattus rattus)22,27,30,31; and S. ratti from the oral swab of a rat32. In two recent animal microbiome projects, other potentially novel Streptobacillus species with ≥91.5% partial 16S rRNA gene sequence identities to known Streptobacillus species were also found in the oral cavities of a Cape ground squirrel (Xerus inauris) (accession number: HM590422, unpublished data) and dogs (Canis familiaris) (accession numbers: EU082091 and JN713542)33,34. In human, the two anatomical sites with the largest number of novel bacteria discovered in the recent two decades are the gastrointestinal tract and oral cavity29. For example, Streptococcus sinensis, a bacterium discovered in 200235 and associated with infective endocarditis in patients from different parts of the world35,36,37,38, was finally also found to be part of human oral flora39. This indicates that there should be yet numerous novel bacterial species remained undescribed from the oral cavity. We speculate that the oral cavities and oropharynx of other animals might be the reservoirs of their own Streptobacillus species. We anticipate that more novel Streptobacillus species will be described and more potentially novel Streptobacillus species will be revealed by molecular epidemiology and microbiome studies in the next decade.
Materials and Methods
Antimicrobial susceptibility
The in vitro antimicrobial susceptibilities of S. hongkongensis against sulfamethoxazole/trimethoprim (range: 1.5625/0.3125 to 400/80 μg/ml) and nalidixic acid (range: 0.15625 to 40 μg/ml) were determined to test for the enrichment condition. Briefly, drug powders of sulfamethoxazole (Fluka, Switzerland), trimethoprim (Fluka), and nalidixic acid (Sigma-Aldrich, St. Louis, MO, USA) were dissolved in sterile dimethyl sulfoxide (Sigma-Aldrich) for the preparation of stock solutions. The stock solutions were then added into 5 ml of BHI broth (Oxoid, UK) to give a series of culture media supplemented with antimicrobial drugs at concentrations within the test range. Approximately 2.5 × 106cfu of S. hongkongensis HKU33T was then added into each of the test medium (final cell density = 5 × 105cfu/ml) and the inoculated broths were incubated under an aerobic environment supplemented with 5% CO2 at 37 °C for 3 days. The presence of growth at each drug concentration was then recorded. The test was also performed using another S. hongkongensis reference strain, HKU34.
Primer design for PCR detection
A pair of primers [LPW21593 (5′-TAGGCGGTTAAACAAGTCAGG-3′) and LPW21594 (5′-TGAGATTCGCTCCATATCGCTATTTCG-3′)] (Invitrogen, Carlsbad, CA, USA) were designed by selecting the conserved regions of the 16S rRNA genes of S. hongkongensis HKU33T and HKU34 (Fig. 3). The specificity of this primer pair was tested using the genomic DNA extracted from strains HKU33T and HKU34, as well as the closely related reference strains S. moniliformis CCUG 13453T, Sneathia sanguinegens CCUG 41628T, and “Sneathia amnii” CCUG 52976, which were obtained from Culture Collection, University of Göteborg (CCUG), Sweden. Bacterial DNA was extracted using the alkaline lysis method. Briefly, cells from a single colony were resuspended in 200 μl of phosphate-buffered saline (PBS) (Oxoid), and 800 μl of NaOH (0.05 mol/L) (Sigma-Aldrich) was added into the bacterial suspensions. The cell suspension mixtures were then heated at 60 °C for 45 min. The lysed cell suspensions were then each neutralized by an addition of 240 μl of Tris-hydrochloric acid (pH 7.0) (Sigma-Aldrich). The neutralized suspensions were then centrifuged at 16,100 rcf for 5 min to remove the cell debris and the total bacterial DNA contained in the supernatant was collected and stored at −20 °C until use.
The nucleotide positions of the primer regions are shown with respect to the complete 16S rRNA gene of S. hongkongensis HKU33T (DDBJ/ENA/GenBank accession number NR_126239).
PCR amplification of the partial 16S rRNA gene was performed using the primer pair LPW21593/LPW21594. Briefly, each 25 μl-PCR reaction mixture contained UltraPure DNase/RNase-free distilled water (Invitrogen, Carlsbad, CA, USA), 1 μl of bacterial DNA, PCR buffer (10 mM of Tris-HCl [pH 8.3], 50 mM of KCl and 2 mM of MgCl2) (Applied Biosystem, Foster City, CA, USA), 1 mM of each primer, 200 μM of each dNTP (Roche Diagnostics, Switzerland), and 0.625 U of AmpliTaq Gold DNA polymerase (Applied Biosystem). The PCR reaction mixtures were first heated at 95 °C for 10 min, then heated in 45 cycles of 95 °C for 1 min, 52.5 °C for 1 min, and 72 °C for 1 min, and finally incubated at 72 °C for 10 min in the GeneAmp PCR System 9700 automated thermal cycler (Applied Biosystem). Five microliters of each amplified product were electrophoresed alongside with the DNA marker GeneRuler 100 bp Plus (Thermo Scientific, Waltham, MA, USA) in 1.5% (w/v) agarose gel (SeaKem LE Agarose) (Lonza, Switzerland) and after electrophoresis the agarose gel was stained with ethidium bromide (Sigma-Aldrich) for DNA band visualization.
Ethics statement
This study has been approved by the Institutional Review Board (IRB) of the University of Hong Kong/Hospital Authority Hong Kong West Cluster and carried out according to the approved protocol. For the use of archived, left over patient throat swab specimens, the requirement of obtaining written informed consent from the patients was waived by the IRB; and no additional patient sample was collected. For the use of control samples, written informed consent was obtained from all the control subjects or their parents or legal guardians.
Patients and controls
All throat swab samples from patients with acute pharyngitis sent to the clinical microbiology laboratory of Queen Mary Hospital, Hong Kong, during a three-month period (1 April 2015 to 30 June 2015) were included in the study. To determine if S. hongkongensis is associated with acute pharyngitis, controls without acute pharyngitis were recruited from healthy volunteers and out-patients. Subjects were excluded if they had immunocompromising conditions including primary immunodeficiency, systemic or inhalational steroid, other immunosuppressive drugs, chemotherapy, diabetes mellitus, chronic organ failure, thalassemia major with regular transfusions and/or iron overload, and hematopoietic stem cell or solid organ transplantation. Subjects who received antibiotics in the preceding 2 weeks prior to throat swabs collection were also excluded. All throat swabs were processed immediately after their arrival at the laboratory.
Bacterial culture
Throat swabs were suspended in BHI broth supplemented with 400/80 μg/ml of sulfamethoxazole/trimethoprim and 40 μg/ml of nalidixic acid. The inoculated broths were then incubated under aerobic environment supplemented with 5% CO2 at 37 °C for 3–5 days.
DNA extraction, PCR, cloning, and DNA sequencing
In order to extract total bacterial DNA from the broth cultures, cells in the broth suspensions were collected by centrifugation at 16,100 rcf for 10 min. Then, the cells were resuspended in 200 μl of phosphate-buffered saline (PBS), and DNA was extracted using the alkaline lysis method as described above. PCR of the partial 16S rRNA gene was performed using the primer pair LPW21593/LPW21594 as described above. DNA from S. hongkonensis HKU33T and autoclaved distilled water were used as the positive and negative controls in each run of PCR, respectively. Preparation of PCR master mix, addition of DNA samples into the reaction tubes and post-PCR steps, including gel electrophoresis, purification of PCR products and DNA sequencing, were performed in separated rooms to avoid possible contamination. After agarose gel electrophoresis, the PCR products were purified using the QIAquick Gel Extraction Kit (QIAGEN) according to the manufacturer’s protocol. Both strands of the PCR products were sequenced twice with an ABI Prism 3700 DNA Analyzer (Applied Biosystems), using the respective PCR primers. The sequencing electropherograms obtained were viewed using Chromas Lite 2.1.1 (Technelysium, Australia).
When ambiguous peaks were observed in the sequencing electropherograms, the PCR products obtained were cloned into plasmids for another trial of sequencing, which was performed according to our previous publication40. Briefly, freshly-prepared gel-purified PCR products were cloned into pCRII-TOPO vector using the TOPO TA Cloning Kit (Invitrogen) according to the manufacturer’s protocol. The TA-ligated plasmids were then transformed into Escherichia coli DH5α (TaKaRa Bio, Japan) by electroporation. The electroporated cells were then grown on LB agar, Lennox (Difco, BD Diagnostic Systems, Sparks, MD, USA) with kanamycin (50 μg/ml) (Sigma-Aldrich) for the selection of positive transformants, as well as isopropyl β-D-1-thiogalactopyranoside (IPTG) (40 μg/ml) (Sigma-Aldrich) and 5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside (X-gal) (100 μg/ml) (Promega, Madison, WI, USA) for blue-white screening. White colonies were then selected and grown in LB broth, Lennox (Difco) with kanamycin (50 μg/ml) overnight and the plasmids in the bacterial cells were extracted using the QIAprep Spin Miniprep Kit (QIAGEN) according to the manufacturer’s protocol. The purified plasmids were then sequenced using the PCR primers directly. The sequencing electropherograms obtained were viewed using Chromas Lite 2.1.1.
Comparative sequence identity analyses and phylogenetic analyses
The sequences obtained from the cloned PCR products were analyzed by pairwise alignment, with the optimal GLOBAL alignment parameters, using BioEdit 7.2.041. The sequences of the PCR products were also compared with sequences of closely related species from the DDBJ/ENA/GenBank databases by multiple sequence alignment using MUSCLE 3.842 and were then end-trimmed. Poorly aligned or divergent regions of the aligned, end-trimmed DNA sequences were removed using Gblocks 0.91b43 with relaxed parameters, and 653 nucleotide positions were included in the final sequence alignment dataset. Tests for substitution models and phylogenetic tree construction using the maximum likelihood method were performed using MEGA 6.0644.
Nucleotide sequence accession numbers
The 16S rRNA gene sequences of the five S. hongkongensis sequence variants detected have been deposited in the DDBJ/ENA/GenBank databases with the accession numbers LC097058- LC097062.
Additional Information
How to cite this article: Lau, S. K. P. et al. Human oropharynx as natural reservoir of Streptobacillus hongkongensis. Sci. Rep. 6, 24419; doi: 10.1038/srep24419 (2016).
References
Schottmüller, H. Zur atiologie und klinik der bisskrankheit (ratten-, katzen-, eichhornchen-bisskrankheit). Dermatol. Wochenschr. 58, 77 (1914).
Levaditi, C., Nicolau, S. & Poincloux, P. Sur le rôle étiologique de Streptobacillus moniliformis (nov. spec.) dans l'érythème polymorphe aigu septicémique. C. R. Acad. Sci. 180, 1188–1190 (1925).
Blake, F. G. The etiology of rat-bite fever. J. Exp. Med. 23, 39–60 (1916).
Faro, S., Walker, C. & Pierson, R. L. Amnionitis with intact amniotic membranes involving Streptobacillus moniliformis . Obstet. Gynecol. 55, 9S–11S (1980).
Torres, L. et al. Bacteremia by Streptobacillus moniliformis: first case described in Spain. Eur. J. Clin. Microbiol. Infect. Dis. 22, 258–260 (2003).
Nei, T. et al. Streptobacillus moniliformis bacteremia in a rheumatoid arthritis patient without a rat bite: a case report. BMC Res. Notes 8, 694 (2015).
Okamori, S. et al. A Japanese patient with a rare case of Streptobacillus moniliformis bacteremia. J Infect. Chemother. 21, 877–878 (2015).
Dijkmans, B. A. C., Mattie, H., Thomeer, R. T. W. M., Vielvoye, G. J. & Lampe, A. S. Brain abscess due to Streptobacillus moniliformis and Actinobacterium meyerii . Infection 12, 262–264 (1984).
Vasseur, E., Joly, P., Nouvellon, M., Laplagne, A. & Lauret, P. Cutaneous abscess: a rare complication of Streptobacillus moniliformis infection. Br. J. Dermatol. 129, 95–96 (1993).
Hagelskjaer, L., Sørensen, I. & Randers, E. Streptobacillus moniliformis infection: 2 cases and a literature review. Scand. J. Infect. Dis. 30, 309–311 (1998).
Madhubashini, M., George, S. & Chandrasekaran, S. Streptobacillus moniliformis endocarditis: case report and review of literature. Indian Heart J. 65, 442–446 (2013).
Fenn, D. W., Ramoutar, A., Jacob, G. & Bin Xiao, H. An unusual tale of rat-bite fever endocarditis. BMJ Case Rep. 10.1136/bcr-2014-204989 (2014).
Pins, M. R., Holden, J. M., Yang, J. M., Madoff, S. & Ferraro, M. J. Isolation of presumptive Streptobacillus moniliformis from abscesses associated with the female genital tract. Clin. Infect. Dis. 22, 471–476 (1996).
Danion, F., Bui, E., Riegel, P. & Goichot, B. Streptobacillosis characterised by palmoplantar pustulosis. Lancet Infect. Dis. 13, 96 (2013).
Wang, T. & Wong, S. Streptobacillus moniliformis septic arthritis: a clinical entity distinct from rat-bite fever? BMC Infect. Dis. 7, 56 (2007).
Flannery, D. D., Akinboyo, I., Ty, J. M., Averill, L. W. & Freedman, A. Septic arthritis and concern for osteomyelitis in a child with rat bite fever. J. Clin. Microbiol. 51, 1987–1989 (2013).
Budair, B., Goswami, K. & Dhukaram, V. Septic arthritis secondary to rat bite fever: a challenging diagnostic course. BMJ Case Rep. 2014, 10.1136/bcr-2014-204086 (2014).
Addidle, M., Pynn, J., Grimwade, K. & Giola, M. Epidural abscess caused by Streptobacillus moniliformis . J. Clin. Microbiol. 50, 3122–3124 (2012).
Chulay, J. D. & Lankerani, M. R. Splenic abscess: report of 10 cases and review of the literature. Am. J. Med. 61, 513–522 (1976).
Dubois, D. et al. Streptobacillus moniliformis as the causative agent in spondylodiscitis and psoas abscess after rooster scratches. J. Clin. Microbiol. 46, 2820–2821 (2008).
Torres, A. et al. Remitting seronegative symmetrical synovitis with pitting edema associated with subcutaneous Streptobacillus moniliformis abscess. J. Rheumatol. 28, 1696–1698 (2001).
Kimura, M. et al. Detection of Streptobacillus spp. in feral rats by specific polymerase chain reaction. Microbiol. Immunol. 52, 9–15 (2008).
Strangeways, W. I. Rats as carriers of Streptobacillus moniliformis . J. Pathol. Bacteriol. 37, 45–51 (1933).
Paegle, R. D., Tewari, R. P., Bernhard, W. N. & Peters, E. Microbial flora of the larynx, trachea, and large intestine of the rat after long-term inhalation of 100 per cent oxygen. Anesthesiology 44, 287–290 (1976).
Woo, P. C. Y. et al. Streptobacillus hongkongensis sp. nov., isolated from patients with quinsy and septic arthritis, and emended descriptions of the genus Streptobacillus and Streptobacillus moniliformis . Int. J. Syst. Evol. Microbiol. 64, 3034–3039 (2014).
Greenwood, J. R. & Harvey, S. M. Streptobacillus moniliformis In The Prokaryotes (eds Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. ) 983–985 (Springer, 2006).
Eisenberg, T. et al. sp. nov. isolated from an Australian spinifex hopping mouse (Notomys alexis) THOMAS, 1922 and emended description of Streptobacillus Levaditi et al. 1925, Eisenberg et al. 2015 emend. Int. J. Syst. Evol. Microbiol. In Press, 10.1099/ijsem.0.000654 (2015).
Eisenberg, T. et al. Streptobacillus felis sp. nov., isolated from a cat with pneumonia, and emended descriptions of the genus Streptobacillus and of Streptobacillus moniliformis . Int. J. Syst. Evol. Microbiol. 65, 2172–2178 (2015).
Woo, P. C. Y., Lau, S. K. P., Teng, J. L. L., Tse, H. & Yuen, K. Y. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 14, 908–934 (2008).
Hopkinson, W. I. & Lloyd, J. M. Streptobacillus moniliformis septicaemia in spinifex hopping mice (Notomys alexis). Aust. Vet. J. 57, 533–534 (1981).
Eisenberg, T. et al. Phenotypic and genotypic characteristics of members of the genus Streptobacillus . Plos One 10, e0134312 (2015).
Eisenberg, T. et al. Streptobacillus ratti sp. nov. isolated from a black rat (Rattus rattus). Int. J. Syst. Evol. Microbiol. In Press, 10.1099/ijsem.0.000869 (2015).
Wouters, E. G. H., Ho, H. T. K., Lipman, L. J. A. & Gaastra, W. Dogs as vectors of Streptobacillus moniliformis infection? Vet. Microbiol. 128, 419–422 (2008).
Dewhirst, F. E. et al. The canine oral microbiome. Plos one 7, e36067 (2012).
Woo, P. C. Y. et al. Streptococcus sinensis sp. nov., a novel species isolated from a patient with infective endocarditis. J. Clin. Microbiol. 40, 805–810 (2002).
Uçkay, I. et al. Streptococcus sinensis endocarditis outside Hong Kong. Emerg. Infect. Dis. 13, 1250–1252 (2007).
Faibis, F. et al. Streptococcus sinensis: an emerging agent of infective endocarditis. J. Med. Microbiol. 57, 528–531 (2008).
Seta, V., Teicher, E., Fortineau, N., Ladouceur, M. & Lambotte, O. Endocardite infectieuse à Streptococcus sinensis . Med. Mal. Infect. 45, 56–57 (2015).
Woo, P. C. Y. et al. The oral cavity as a natural reservoir for Streptococcus sinensis . Clin. Microbiol. Infect. 14, 1075–1079.
Zhao, Y. et al. Intra-genomic internal transcribed spacer region sequence heterogeneity and molecular diagnosis in clinical microbiology. Int. J. Mol. Sci. 16, 25067–25079 (2015).
Hall, T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 (1999).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Acknowledgements
This work is partly supported by the Strategic Research Theme Fund, Mary Sun Medical Scholarship, Wong Ching Yee Medical Postgraduate Scholarship, and University Postgraduate Scholarship, The University of Hong Kong, Hong Kong; and the Croucher Senior Medical Research Fellowship, Croucher Foundation, Hong Kong.
Author information
Authors and Affiliations
Contributions
S.K.P.L. and P.C.Y.W. conceived the idea and initiated the study. S.K.P.L., J.F.W.C., C.-C.T., S.-M.C. and Y.-L.L. recruited the control subjects and obtained the throat swab samples. C.-C.T. and M.-L.H. performed the experiments. S.K.P.L., J.F.W.C., C.-C.T., S.-M.C., M.-L.H., T.-L.Q., Y.-L.L. and P.C.Y.W. analyzed the data and reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Lau, S., Chan, J., Tsang, CC. et al. Human oropharynx as natural reservoir of Streptobacillus hongkongensis. Sci Rep 6, 24419 (2016). https://doi.org/10.1038/srep24419
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep24419
- Springer Nature Limited
This article is cited by
-
Mucosa‐associated cultivable aerobic gut bacterial microbiota among colorectal cancer patients attending at the referral hospitals of Amhara Regional State, Ethiopia
Gut Pathogens (2021)
-
Revealing oral microbiota composition and functionality associated with heavy cigarette smoking
Journal of Translational Medicine (2020)
-
Streptobacillus felis, a member of the oropharynx microbiota of the Felidae, isolated from a tropical rusty-spotted cat
Antonie van Leeuwenhoek (2020)