Abstract
The relationships between polymorphisms of the trans-membrane(TM) region located in the major histocompatibility complex (MHC) class I chain–related gene A (MICA) and Behcet’s disease (BD) have been discussed previously, however, the results were contradictory. In this study, we thoroughly assess whether MICA-TM gene variants are associated with BD by means of a systematic review and meta-analysis. Our study focused on the effects of polymorphisms of MICA-A4, A5, A5.1, A6 and A9 from the included articles. Sixteen previous original publications representing 1,555 BD patients and 2,086 unrelated healthy controls analyzed the association of BD with MICA-TM gene polymorphisms. For the five alleles, MICA-A6 showed a strongly positive correlation with BD patients and could be viewed as an increased risk factor of BD (OR = 2.34, 95%CI: 2.02–2.70). Furthermore, MICA-A4, A5, A5.1 and A9 exhibited negative associations with BD (OR = 0.71, 95%CI: 0.58–0.86; OR = 0.75, 95%CI: 0.63–0.90; OR = 0.63, 95%CI: 0.44–0.91; OR = 0.70, 95%CI: 0.58–0.84, respectively). Our meta-analysis confirmed MICA-A6 could be responsible for BD in three ethnic regions and should probably be treated as a risk factor for BD. MICA-A4, A5, A5.1 and A9 could be regarded as protective factors, especially in the Middle East and East Asia.
Similar content being viewed by others
Introduction
Behcet’s disease (BD) is a refractory multi-system inflammatory disease, characterized by four common manifestations, as follows: recurrent genital ulcerations, oral aphthous ulcers, skin lesions and ocular lesions, along with symptoms in the gastrointestinal tract, central nervous system, vascular system, joints, kidneys and lungs1. It has been observed worldwide in many ethnic groups, but most commonly in patients from Japan, China and Korea, as well as along the Silk Route to the countries of the Mediterranean2. Although its etiology and pathogenesis are still undefined, multiple genetic factors and environmental risk factors such as infectious triggers are considered to confer susceptibility to the disease3.
Many genes have been reported to be associated with BD, including STAT4, interleukin-23 receptor(IL23R), CD40 and IL174. Up to the present, the HLA-B51 molecule has had the strongest known genetic association with BD in many different ethnic groups2,5,6,7. However, whether disease susceptibility is influenced by HLA-B51 itself or by some other genes located around HLA-B in linkage disequilibrium with HLA-B51 remains controversial. Recently, the major histocompatibility complex (MHC) class I chain–related gene A (MICA), a functional gene located between the HLA-B and tumor necrosis factor(TNF) genes on the short arm of human chromosome 6, has been reported to be linked with BD in the trans-membrane (TM) region1,8,9. As a stress-inducible antigen, MICA plays an important role in innate and adaptive immune responses by interacting with the natural killer group 2 member D (NKG2D)-activating receptor of natural killer(NK) cells, CD8 T cells and γδT cells10. Exon 5 in the MICA-TM gene is composed of at least five variable alleles (A4, A5, A5.1, A6 and A9) presenting 4, 5, 6 and 9 triplet repeats of (GCT/AGC)1. Polymorphisms of the TM region have been studied to investigate the association with BD in several articles, but the results are still disputed, probably due to the different ethnicities, smaller sample size and bias in the chosen patients or controls in these works.
In order to better understand the genetic risk of MICA-TM in the relationship with BD, we performed a systematic review and meta-analysis to illuminate this association and determine whether the polymorphisms of MICA-TM conferred susceptibility to BD.
Results
General characteristics of studies
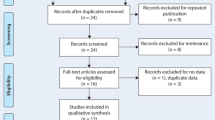
The selection process of eligible studies is shown in Fig. 1A total of 202 reports were obtained from PubMed, Embase, Web of Science, CBM and CNKI. Among them, 175 articles were excluded; of which 115 were duplicates and 60 were reviews, meeting reports or articles unrelated to the topic. The other 27 articles were all full-text. Eight articles were excluded because they did not discuss MICA-TM allele variants11,12,13,14,15,16,17,18, two were excluded because they only compared controls19,20 and one was excluded because of overlapping patient information with another paper8. Finally, 16 publications concerning about the distribution of MICA-TM gene were available for our topic in this meta-analysis1,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35. The characteristics of the selected studies are presented in Table 1.
Two articles1,24 written by the same author had an overlapping sample from Japan and one article1 had more detailed genotyping data, so we decided to exclude the case-control group from Japan in the second article24. Therefore, we conducted a meta-analysis that combined 13 studies from 16 articles including 1,304 BD patients and 1,826 unrelated healthy controls for MICA-A4, MICA-A5.1 and MICA-A9; 14 studies including 1,353 BD patients and 1,876 controls for MICA-A5; and 16 studies involving 1,555 BD patients and 2,086 controls for MICA-A6.
Bias assessment of the included studies
Potential bias assessment of the included studies is presented in Table 2. Among the studies, three articles(18.8%)1,21,28 had bias of ascertainment in the selection of cases, two (12.5%)30,33 showed possible bias in the population stratification and the others demonstrated no bias in the selection of cases, controls, genotyping controls, confounding variables, multiple tests, or selective outcome reports.
Association between MICA-TM allele polymorphisms and BD susceptibility
Sixteen articles on three ethnicities—namely Middle Eastern, Caucasian and East Asian populations—were incorporated in our meta-analysis. The association between MICA-A4, A5, A5.1, A6, A9 and BD susceptibility was analyzed (Table 3, Fig. 2). We first used STATA 12.0 software to investigate the pooled ORs and heterogeneity. Among the five allele polymorphisms of MICA-TM, the allele MICA-A4, which was studied in 13 studies, was found to be associated with BD. Although only one OR was statistically significant in the relationship with BD, the pooled OR was 0.71(95%CI: 0.58–0.86, Fig. 2A). The MICA-A5 allele was investigated in 14 studies and only two of the all research had a positive relationship with BD. The pooled OR was 0.75(95%CI: 0.63–0.90, Fig. 2B). The pooled OR of the MICA-A5.1 allele from 13 studies was 0.63(95%CI: 0.44–0.91, Fig. 2C). The MICA-A6 allele was investigated from 18 studies and 11 of these were related to BD. The pooled OR was 2.34(95%CI: 2.02–2.70, Fig. 2D). The MICA-A9allele, which was reported to show a statistically significant difference in one article, demonstrated significance and the pooled OR was 0.70(95%CI: 0.58–0.84, Fig. 2E).
We then performed a sub-group analysis based on ethnicity. For the MICA-A4allele, statistics derived from three ethnicities (Middle Eastern, Caucasian and East Asian) indicated an overall I2 of 0.0% and ORs of 0.69(95%CI: 0.50–0.94), 0.81(95%CI: 0.56–1.18) and 0.66(95%CI: 0.47–0.93), respectively. For the MICA-A5allele, statistics derived from the three ethnicities showed an overall I2 of 0.6% and ORs of 0.61(95%CI: 0.45–0.82), 0.96(95%CI: 0.64–1.45) and 0.80(95%CI: 0.62–1.04), respectively.
For the MICA-A5.1allele, statistics derived from the three ethnicities showed an overall I2 of 63.0% and ORs of 0.69(95%CI: 0.51–0.92), 0.74(95%CI: 0.38–1.44) and 0.40(95%CI: 0.19–0.83), respectively. For the MICA-A6allele, statistics derived from the three ethnicities showed an overall I2 of 25.0% and ORs of 2.32(95%CI: 1.88–2.87), 2.13(95%CI: 1.57–2.89) and 2.54(95%CI: 1.95–3.31), respectively. Finally, for MICA-A9, the statistics derived from the three ethnicities showed an overall I2 of 0.0% and ORs of 0.72(95%CI: 0.54–0.97), 0.59(95%CI: 0.40–0.86) and 0.77(95%CI: 0.55–1.06), respectively.
Sensitivity analysis
Sensitivity analysis was conducted by the authors to evaluate the effect of each study on the pooled ORs by omitting each study in turn. The pooled ORs were not affected by excluding any study (data not shown).
Publication bias
A series of Begg’s funnel plots and Egger’s regression tests were applied to detect the publication bias for the five alleles of MICA-TM. Then, five symmetrical funnel plots were depicted for all of the alleles (data not shown). As shown in Table 3, we did not find any obvious publication bias for the association between MICA-TM and BD.
Discussion
Articles referring to the relationship between MICA-TM polymorphisms and BD have been published over the past 10 years. In this meta-analysis, we included 1,555 BD patients and 2,086 unrelated healthy controls from 16 articles and performed a detailed review related to three ethnicities. The results suggested that the MICA-A6 allele can be treated as an increased risk factor of BD, with a pooled OR of 2.34 while the MICA-A4, A5, A5.1 and A9 alleles can be cautiously viewed as protective factors, with ORs of 0.71, 0.75, 0.63 and 0.70, respectively. Importantly, the MICA-A6 allele was associated with BD in all three ethnic groups. The other alleles were only relevant in Middle Eastern or East Asian patients with BD. In other words, the MICA-A4, A5, A5.1 and A9 allele polymorphism analyzed was not significantly associated with BD in patients from non–Silk Route regions. It could be that the relatively small number of subjects in each study can explain this inconsistent result.
The MICA gene is the nearest neighbor of HLA-B identified to data(only 47kb centromeric) and is by far the most divergent MHC-I known. Similar to the protein fold of MHC class I and homologs, the structure of MICA gene contains long open-reading frames encoding for MHC class I molecules with three distinct extracellular domains(α1, α2 and α3), a transmembrane segment and a cytoplasmic tail, each encoded by a separate exon36. Steinle et al.37 found that a single amino acid substitution at position 129 in the α2 domain of MICA altered the affinity of binding to the activating natural killer group 2, member D (NKG2D). Zou et al.38 reported that a nucleotide insertion at the transmembrane region of MICA, resulting a truncated TM region, lead to resist its down-regulation and thereby is functionally relevant in the elimination of virus-infected cells. Whether external region or TM region is more important for the functional role of MICA remains unclear. Further studies are needed to elucidate the exact roles of MICA.
As with classical MHC-I gene, MICA is characterized by its high degree of allele polymorphism which are mostly localized in the exon 2, 3 and 4 (extracellular domains)39. Steinle et al.37 found that MICA*01 and *07 in the α1α2 domain, but not MICA*04, *08 and *16, had a reduced binding affinity with NKG2D and the amino acid substitution of methionine by valine likely affected NKG2D binding indirectly by a conformational change. Additionally, unusual variability in exon 5 presents a microsatellite polymorphism encoding a distinct number of alanine residues in the transmembrane domain corresponding to the microsatellite alleles A4, A5, A5.1, A6, A92. Steinle and co-workers40 believed that these changes did not alter the overall hydrophobic character of the molecule, neither affected surface expression of MICA, so that the function of this variability may be questionable. Even though we still did not know whether the extracellular domains of the MICA molecule or the transmembrane region determined the significant function related to the mechanism of immune response, a possible hypothesis have been proposed that a single amino acid insertion/deletion in the MICA-TM region with α-helix leads a net of rotation of about 100° of the extracellular with respect to the cytoplasmic domain30. Maybe, this would result in substantial modification of the intermolecular interaction mediated by both the extracellular and cytoplasmic parts of MICA. As a result of this assumption, the pronounced difference in binding affinities of triplet repeats for NKG2D could have significant effects on NK cell activation and the modulation of T-cell response,which could play a role in precipitating or exacerbating autoimmune response.
Yabuki and his colleagues23 argued that two hypotheses can account for the primary involvement of the MICA molecule with BD. First, the local immune response in vivo is induced after bacterial infection, resulting in stress-induced expression of MICA. Secondly, some bacterial components could have a specific role similar to that of super-antigens in the activation of the MICA molecule. Both hypotheses indicate that the increased MICA-A6 molecule may activate γδT cells, thereby triggering the unusual immune response related with BD.
Although the role of polymorphisms in the TM region of MICA gene is still under debate, the potential correlation between MICA-TM and the development of BD has been investigated in different ethnic groups, including East Asian, Caucasian and Middle Eastern populations from Japan through to Israel. In our meta-analysis, MICA-A6, the most investigated allele, was found to confer susceptibility to BD with a pooled OR of 2.34 in all three ethnic groups. This result is consistent with those of the original articles included in our study, suggesting that A6 was a common risk gene for BD. Additionally, this gene was a causative risk gene or strongly linked with the true risk gene of BD. The functional or fine-mapping studies are needed to elucidate the exact role of A6 or this region and will helpful for common drug discovery of BD suitable for all ethnic populations. Picco and his colleagues30 found that secreted MICA-A6 may provide better steric conditions for ligation, such as bacterial component binding with γδT cells and NK cells that express MICA molecules, thus leading to the onset of BD. The functional correlation between MICA-A4, A5, A5.1 and A9 and BD has not been reported previously. Only a few of our included articles indicated the association of BD with MICA-A4, A5, A5.1 and A9.
The TM region contains most of the hydrophobic amino acids and is mainly expressed in epithelial cells, fibroblasts, endothelial cells and monocytes3,41. Nishiyama and his colleagues25 suggested that the A4 allele has a high negative correlation with ocular lesions and the A5 allele has a negative relationship with iridocyclitis in BD patients. Picco and colleagues30 argued that MICA-A5.1 seems to play a protective role in BD patients. Furthermore, Park and colleagues29 suggested that patients with allele A9 have less severe BD complications than those without allele A9 in terms of uveitis, thrombosis and neurological and intestinal involvement.
The common opinion that BD shows a strong association with HLA-B51 has been disclosed in relation to several ethnic groups, including East Asian, Caucasian and Middle Eastern populations. A previous study32, together with results related to Spanish22, Greek23 and Italian28 populations, as well as results presented by Mizuki and his colleagues24, indicated that the MICA-TM molecule is strongly associated with BD owing to the linkage disequilibrium with HLA-B51. Additionally, previous studies also showed that MICA-A6 is linked with HLA-B52. However, HLA-B52 didn’t associate with Behcet’s disease. These may be explained by the different linkage models in patients and controls for between MICA and HLA-B52. MICA-TM (A9) was found in linkage disequilibrium with HLA-B52 in controls but not in patients with BD21. Many BD patients are HLA-B51 negative, but with another Bw4 allele, which is not associated with BD, suggesting that in addition to HLA-B51, there are other gene play important roles in the development of BD.Mizuki and his colleagues1 and Park et al.29 suggested that MICA-TM alleles rather than HLA-B51 play an important role in the development of BD. Especially for HLA-B51-negative patients from Korea, MICA-A6 could be viewed as a meaningful susceptibility biomarker.
A recent study investigated by Hughes et al.18 suggested that a noncoding variant site(rs116799036), between the HLA-B and MICA gene, was the true source of BD risk factor. This implied that the risk generally ascribed to HLA-B51 was likely not causal in BD. Conversely, the other study reported by Ombrello et al.42 indicated that HLA-B51 was much more strongly associated with BD than any SNP and conferred significant risk for BD even after controlling for the effect of rs116799036. These contrary results may be explained by population heterogeneity or statistical methodologies. Hughes et al.18 performed the HLA genotyping at Turkish and Italian populations and used a reference panel of Northern European ancestry for HLA imputation, whereas Ombrello et al.43 examined HLA genotyping at Turkish population and used a reference panel of mixed European ancestry.
Collectively, the MICA-TM gene appears to be a strong candidate gene for BD based on three main aspects, as follows: its chromosomal localization23, its restricted and heat shock–induced expression in epithelial cells44 and its predicted immunological function as a ligand of NK cells and γδT cells10. Although it has not been determined whether the HLA-B51 gene itself or the nearby MICA-TM gene B is directly localized in the pathogenesis of BD, the possibility must exist that susceptibility or co-susceptibility gene(s) within the genomic sequence region could be implicated in association with BD. Thus, tests with a combination of HLA-B51 and MICA-TM may act as a better genetic marker for BD.
Despite considerable efforts to detect the potential relationship between MICA-TM alleles and BD, some limitations of this meta-analysis need to be mentioned. First, heterogeneity among the ethnic groups was discovered when investigating the association of MICA-TM with BD. However, based on the results of the sensitivity analysis, it is clear that the overall effect was not affected by heterogeneity. Second, the number of patients and controls was relatively small in each included study; therefore, a much larger sample size from different ethnic populations is required for further analysis. Third, since the ethnic origins of patients and controls were not specified in any of the studies, subjects’ ethnicity and different criteria for controls are potential sources of heterogeneity. Finally, the databases from which we selected eligible studies were English and Chinese; thus, a language bias may have been present in our meta-analysis.
In conclusion, our results demonstrated that MICA-A6 probably confers a strong susceptibility to BD in three ethnic regions and could be treated as a risk factor for BD. MICA-A4, A5, A5.1 and A9 could be regarded as protective factors, especially in the Middle East and East Asia. However, these relationships need to be demonstrated from a pathogenic point of view.
Materials and Methods
Search strategy
Articles were collected from the following electronic databases: PubMed, Embase, Web of Science, the China Biomedical (CBM) database and the China National Knowledge Infrastructure (CNKI) database. All studies were carefully selected and were up to date as of May 17, 2015. The following subject headings and key words were used: “Behcet syndrome,” “Behcet’s syndrome,” “Behcets syndrome,” “Behcet disease,” “Behcet’s disease,” or “BD” and “MHC class I chain–related gene A,” “MICA,” or “MIC-A”, without any limitation imposed.
Study selection
The retrieved articles selected from electronic databases were archived by two reviewers independently by inspecting the title, abstract and full-text according to specified standards. Any discordance could be solved through discussion and consensus in collaboration with a third author. Included studies in this meta-analysis needed to meet the following criteria: 1) they sought to determine the association between MICA-TM and BD; 2) a detailed number or percent of MICA-TM alleles could be obtained for cases and controls; 3) they were focused on human beings; and 4) they used the case-control approach. The exclusion criteria were as follows: 1) duplication of a previous article; 2) studies that were case reports, reviews or letters; and 3) insufficient data was provided after contacting the corresponding author.
Data extraction
Data from the selected studies were extracted independently by two reviewers (J.Z. and D.L.). The following contents from each study were collected: name of first author, year of publication, country of cases and controls, ethnicity, characteristics and number of cases and controls, genotyping method, diagnostic criteria for BD and frequency or percentage of MICA-TM alleles in cases and controls. Two authors carefully checked the collected data and reached agreement on all decisions. For any disagreements that still existed, a third investigator was asked to resolve the issue through discussion.
Quality assessment
Quality evaluation of the extracted studies was also performed by two authors (J.Z. and D.L.) based on the HuGENetHandbook45. Six bias assessment items referring to gene-disease association were incorporated in this handbook, including bias in selection of cases, bias in selection of controls, bias in genotyping cases, bias in genotyping controls, bias in population stratification, confounding bias, multiple tests and selective outcome reports. The quality ascertainment of every item ranged from “Yes” to “No,” while the label “Unclear” was used if there was not enough information to make a determination. A correction and review was carried out by another author (L.Y.) independently if the two authors dissented with each other’s view. Consensus needed to be attained for all labels after discussion.
Statistical analysis
The systematic checklists and guidelines in the HuGENetHandbook45 were applied to perform this meta-analysis. The odds ratio (OR) and 95% confidence interval (CI) were calculated and pooled ORs were analyzed for MICA-TM frequency comparison between BD patients and controls. Cochran’s Q statistic was used to assess heterogeneity (p < 0.1, treated as significant level across studies). Moreover, the quantitative I2 statistic was used for estimation of inconsistency in our meta-analysis, representing the percentage of the observed variability due to heterogeneity rather than to chance (no heterogeneity, I2 = 0–25%; moderate heterogeneity, I2 = 25–50%; largeheterogeneity, I2 = 50–75%; extreme heterogeneity, I2 = 75–100%)46. Either the fixed-effect model (I2 < 25% and p > 0.1) or random-effect model (I2 ≥ 25% and p < 0.1) was applied for the pooled ORs and 95%CIs according to the heterogeneity. We conducted sensitivity analysis to assess the effect of each study on the pooled ORs by omitting each study in turn. Moreover, subgroup analysis was performed to determine the strength of association of different ethnicities. Publication bias was also checked by Begg’s funnel plots47 and Egger’s regression test48. STATA12.0 software (StataCorp LP, College Station, Texas, USA) was used to carry out statistical analysis. A significant difference was estimated under the level of 0.05 (a two-tailed p value) except for the Q statistic. All results had to be validated by two authors (J.Z. and D.L.) independently.
Additional Information
How to cite this article: Zhang, J. et al. Association between Functional MICA-TM and Behcet’s Disease: A Systematic Review and Meta-analysis. Sci. Rep. 6, 21033; doi: 10.1038/srep21033 (2016).
References
Mizuki, N. et al. Triplet repeat polymorphism in the transmembrane region of the MICA gene: a strong association of six GCT repetitions with Behcet disease. Proc Natl Acad Sci USA 94, 1298–1303 (1997).
Ohno, S. et al. Close association of HLA-Bw51 with Behcet’s disease. Arch Ophthalmol 100, 1455–1458 (1982).
Bahram, S., Bresnahan, M., Geraghty, D. E. & Spies, T. A second lineage of mammalian major histocompatibility complex class I genes. Proc Natl Acad Sci USA 91, 6259–6263 (1994).
Hou, S., Kijlstra, A. & Yang, P. The genetics of Behcet’s disease in a Chinese population. Front Med 6, 354–359 (2012).
Barnes, C. G. & Yazici, H. Behcet’s syndrome. Rheumatology 38, 1171–1174 (1999).
Sakane, T., Takeno, M., Suzuki, N. & Inaba, G. Behcet’s disease. N Engl J Med 341, 1284–1291 (1999).
Verity, D. H., Marr, J. E., Ohno, S., Wallace, G. R. & Stanford, M. R. Behcet’s disease, the Silk Road and HLA-B51: historical and geographical perspectives. Tissue Antigens 54, 213–220 (1999).
Ota, M. et al. The critical region for Behcet disease in the human major histocompatibility complex is reduced to a 46-kb segment centromeric of HLA-B, by association analysis using refined microsatellite mapping. Am J Hum Genet 64, 1406–1410 (1999).
Mizuki, N., Inoko, H. & Ohno, S. Molecular genetics (HLA) of Behcet’s disease. Yonsei Med J 38, 333–349 (1997).
Groh, V., Steinle, A., Bauer, S. & Spies, T. Recognition of stress-induced MHC molecules by intestinal epithelial gammadelta T cells. Science 279, 1737–1740 (1998).
Kimura, T. et al. Microsatellite polymorphism within the MICB gene among Japanese patients with Behcet’s disease. Hum Immunol 59, 500–502 (1998).
Marin, M. L., Savioli, C. R., Yamamoto, J. H., Kalil, J. & Goldberg, A. C. MICA polymorphism in a sample of the Sao Paulo population, Brazil. Eur J Immunogenet 31, 63–71 (2004).
Verity, D. H. et al. Intercellular adhesion molecule-1 gene polymorphisms in Behcet’s disease. Eur J Immunogenet 27, 73–76 (2000).
Hughes, E. H. et al. Associations of major histocompatibility complex class I chain-related molecule polymorphisms with Behcet’s disease in Caucasian patients. Tissue Antigens 66, 195–199 (2005).
Munoz-Saa, I. et al. Allelic diversity and affinity variants of MICA are imbalanced in Spanish patients with Behcet’s disease. Scand J Immunol 64, 77–82 (2006).
Mizuki, N. et al. Association of Major Histocompatibility Complex Class I Chain-Related Gene A and HLA-B Alleles with Behcet’s Disease in Turkey. Jpn J Ophthalmol 51, 431–436 (2007).
Kim, S. J. et al. Targeted resequencing of candidate genes reveals novel variants associated with severe Behcet’s uveitis. Exp Mol Med 45, e49 (2013).
Hughes, T. et al. Identification of multiple independent susceptibility loci in the HLA region in Behcet’s disease. Nat Genet 45, 319–324 (2013).
Nishiyama, M. et al. Research report: Frequencies of mica gene polymorphism: a comparison between Indonesians on Bacan Island and suburban Japanese. Southeast Asian J Trop Med Public Health 35, 195–201 (2004).
Durmanova, V. et al. Characterization of MICA gene polymorphism of HLA complex in the Slovak population. Ann Hum Biol 38, 570–576 (2011).
Wallace, G. R. et al. MIC-A allele profiles and HLA class I associations in Behcet’s disease. Immunogenetics 49, 613–617 (1999).
Gonzalez-Escribano, M. F. et al. Lack of association of MICA transmembrane region polymorphism and Behcet’s disease in Spain. Tissue Antigens 54, 278–281 (1999).
Yabuki, K. et al. Association of MICA gene and HLA-B*5101 with Behcet’s disease in Greece. Invest Ophthalmol Vis Sci 40, 1921–1926 (1999).
Mizuki, N. et al. Localization of the pathogenic gene of Behcet’s disease by microsatellite analysis of three different populations. Invest Ophthalmol Vis Sci 41, 3702–3708 (2000).
Nishiyama, M. et al. Microsatellite polymorphisms of the MICA gene among Japanese patients with Behcet’s disease. Can J Ophthalmol 41, 210–215 (2006).
Ben Ahmed, M. et al. MICA transmembrane region polymorphism and HLA B51 in Tunisian Behcet’s disease patients. Adv Exp Med Biol 528, 225–228 (2003).
Mizuki, N. et al. Microsatellite mapping of a susceptible locus within the HLA region for Behcet’s disease using Jordanian patients. Hum Immunol 62, 186–190 (2001).
Salvarani, C. et al. Association of MICA alleles and HLA-B51 in Italian patients with Behcet’s disease. J Rheumatol 28, 1867–1870 (2001).
Park, S. H. et al. Association of MICA polymorphism with HLA-B51 and disease severity in Korean patients with Behcet’s disease. J Korean Med Sci 17, 366–370 (2002).
Picco, P. et al. MICA gene polymorphisms in an Italian paediatric series of juvenile Behcet disease. Int J Mol Med 10, 575–578 (2002).
Piga, M. et al. Genetics of Behcet’s disease in Sardinia: two distinct extended HLA haplotypes harbour the B*51 allele in the normal population and in patients. Clin Exp Rheumatol 30, S51–56 (2012).
Cohen, R., Metzger, S., Nahir, M. & Chajek-Shaul, T. Association of the MIC-A gene and HLA-B51 with Behcet’s disease in Arabs and non-Ashkenazi Jews in Israel. Ann Rheum Dis 61, 157–160 (2002).
Mok, J., Bang, D., Lee, E. S., Lee, S. & Park, K. Strong association of MIC-A*009 of extracellular domains and MIC-A*A6 of transmembrane domain in Korean patients with Behcet’s disease. Adv Exp Med Biol 528, 221–224 (2003).
Carapito, R. et al. On the genetics of the Silk Route: association analysis of HLA, IL10 and IL23R-IL12RB2 regions with Behcet’s disease in an Iranian population. Immunogenetics 67, 289–293 (2015).
Mizuki, N. et al. Analysis of microsatellite polymorphism around the HLA-B locus in Iranian patients with Behcet’s disease. Tissue Antigens 60, 396–399 (2002).
Li, P. et al. Complex structure of the activating immunoreceptor NKG2D and its MHC class I-like ligand MICA. Nat Immunol 2, 443–451 (2001).
Steinle, A. et al. Interactions of human NKG2D with its ligands MICA, MICB and homologs of the mouse RAE-1 protein family. Immunogenetics 53, 279–287 (2001).
Zou, Y., Bresnahan, W., Taylor, R. T. & Stastny, P. Effect of human cytomegalovirus on expression of MHC class I-related chains A. J Immunol 174, 3098–3104 (2005).
Perez-Rodriguez, M. et al. Further polymorphism of the MICA gene. Eur J Immunogenet 29, 35–46 (2002).
Steinle, A., Groh, V. & Spies, T. Diversification, expression and gamma delta T cell recognition of evolutionarily distant members of the MIC family of major histocompatibility complex class I-related molecules. Proc Natl Acad Sci USA 95, 12510–12515 (1998).
Zwirner, N. W., Fernandez-Vina, M. A. & Stastny, P. MICA, a new polymorphic HLA-related antigen, is expressed mainly by keratinocytes, endothelial cells and monocytes. Immunogenetics 47, 139–148 (1998).
Ombrello, M. J. et al. Behcet disease-associated MHC class I residues implicate antigen binding and regulation of cell-mediated cytotoxicity. Proc Natl Acad Sci USA 111, 8867–8872 (2014).
Remmers, E. F. et al. Genome-wide association study identifies variants in the MHC class I, IL10 and IL23R-IL12RB2 regions associated with Behcet’s disease. Nat Genet 42, 698–702 (2010).
Groh, V. et al. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci USA 93, 12445–12450 (1996).
Little, J. & Higgins, J. P. T. (editors). The HuGENetTMHuGE Review Handbook, version 1.0. (2006) Available at: http://www.hugenet.ca. (Accessed: 4th April 2015).
Higgins, J. P., Thompson, S. G., Deeks, J. J. & Altman, D. G. Measuring inconsistency in meta-analyses. BMJ 327, 557–560 (2003).
Begg, C. B. & Mazumdar, M. Operating characteristics of a rank correlation test for publication bias. Biometrics 50, 1088–1101 (1994).
Egger, M., Davey Smith, G., Schneider, M. & Minder, C. Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634 (1997).
Acknowledgements
This work was supported by National Natural Science Foundation Project (81522013, 81270990), Chongqing Outstanding Youth Grant (cstc2014jcyjjq10005).
Author information
Authors and Affiliations
Contributions
S.H. designed the study. J.Z., D.L. and L.Y. collected and checked the information of eligible articles included in this meta-analysis. J.Z. and S.H. analyzed the data. J.Z. wrote the main manuscript text. S.H. revised the manuscript. All authors reviewed and approved the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhang, J., Liao, D., Yang, L. et al. Association between Functional MICA-TM and Behcet’s Disease: A Systematic Review and Meta-analysis. Sci Rep 6, 21033 (2016). https://doi.org/10.1038/srep21033
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep21033
- Springer Nature Limited