Abstract
Degenerative aortic stenosis (AS) is the most common worldwide cause of valve replacement. The aortic valve is a thin, complex, layered connective tissue with compartmentalized extracellular matrix (ECM) produced by specialized cell types, which directs blood flow in one direction through the heart. There is evidence suggesting remodeling of such ECM during aortic stenosis development. Thus, a better characterization of the role of ECM proteins in this disease would increase our understanding of the underlying molecular mechanisms. Aortic valve samples were collected from 18 patients which underwent aortic valve replacement (50% males, mean age of 74 years) and 18 normal control valves were obtained from necropsies (40% males, mean age of 69 years). The proteome of the samples was analyzed by 2D-LC MS/MS iTRAQ methodology. The results showed an altered expression of 13 ECM proteins of which 3 (biglycan, periostin, prolargin) were validated by Western blotting and/or SRM analyses. These findings are substantiated by our previous results demonstrating differential ECM protein expression. The present study has demonstrated a differential ECM protein pattern in individuals with AS, therefore supporting previous evidence of a dynamic ECM remodeling in human aortic valves during AS development.
Similar content being viewed by others
Introduction
With a growing older population, cardiovascular diseases are becoming an increasing economic and social burden in Western societies. Cardiovascular calcification is a major characteristic of chronic inflammatory disorders such as chronic renal disease, atherosclerosis and calcific aortic valve disease, which are associated with significant morbidity and mortality1. Currently, degenerative aortic stenosis (AS) is the main cause of aortic valve replacement in developed countries. It is characterized by a narrowing of the aortic valve opening which causes the obstruction of the blood flow from the left ventricle into the aorta2. With all this in mind, it is important to note that the ability of the valve to open and close repeatedly, as well as the maintenance of the phenotypes of valvular cells, is made possible by the tissue microstructure. This microstructure is influenced by the composition and orientation of the extracellular matrix (ECM). The ECM within the aortic valve is a dynamic environment rich in collagen, elastic fibers, proteoglycans and glycosaminoglycans, growth factors, proteases, signaling molecules and secreted proteins3. The ECM provides an organized framework which enforces cell orientation and structural integrity, allowing valve ability. Indeed, ECM is involved in valvular remodeling, which is typically described as a process composed of three main overlapping phases: inflammatory, proliferative and maturation4,5. Despite the growing evidence connecting ECM remodeling with AS pathogenesis, the underlying regulatory mechanisms for such remodeling and stratification are not well defined, even though they seem relevant to interpret aortic valve degeneration. In previous studies in our lab6, we conducted an in-depth analysis of the secretome of aortic valves which pointed out an outstanding role of valvular remodeling in the pathophysiological mechanisms of AS. The functional classification of the differentially secreted proteins showed a significant number of ECM proteins. Since many ECM proteins are involved in AS and taking into account its high prevalence and the long course of this disease, a better understanding of AS at the molecular level may be possible through a proteomic approach. This powerful technique might provide useful information to better understand the development of the disease. Besides, it may offer a new gate for the discovery of novel potential biomarkers shed to blood by the aortic valve, which may allow detecting patients at risk of developing degenerative AS, as a starting point.
In fact, previous studies of our group using two-dimensional difference in-gel electrophoresis (2D-DIGE) showed several ECM proteins altered with the progression of AS.6,7. According to our background, the aim of this work is to further clarify AS process and to ascertain whether ECM proteins might play a determinant role in AS. Here, we compared human AS valve tissue with that of healthy subjects using isobaric tags for relative and absolute quantitation (iTRAQ), a robust technique which presents more sensitivity and better accuracy of quantification than 2D-DIGE8 aiming to complement previous analyses with AS tissue using a deeper approach. Validation of 3 ECM proteins found altered was performed by orthogonal techniques (Western blotting and/or selected reaction monitoring (SRM)).
Results
Quantitative proteomics: iTRAQ labeling
To provide insight into the consequences of AS on protein expression, we compared the proteome of AS tissue with the one from healthy valves. Lysates from 4 control samples and 4 stenotic samples were iTRAQ labeled as shown in Fig. 1 of supplementary material (Figure SM1). Samples were pooled in four labeled peptide mixtures to allow the analysis of all of them in a limited number of LC-MS/MS runs avoiding labeling bias (for experimental design see supplementary material). After the labeling procedure and liquid chromatography (LC) separation, derived peptides were identified and quantified by tandem mass spectrometry (MS/MS). The data generated by liquid chromatography coupled to mass spectrometry (LC-MS/MS) analysis were searched against the human RefSeq database using Proteome Discoverer software (Thermo Scientific).
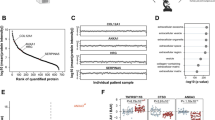
Schematic representation of the proteins identified in the iTRAQ experiment.
Within the 712 identified proteins, 56 proteins (34 up-regulated and 19 down-regulated in stenotic aortic valves when compared to controls, in addition to 3 proteins which are increased or decreased depending on the isoform identified in stenotic valves when compared to controls) were detected in the four runs (mixtures) of the experiment carried out.
iTRAQ ratios corresponding to 712 proteins between control and stenotic samples in four separate experiments were compared (Fig. 1).
For each experiment, we examined the original protein list generated by Protein Discoverer and filtered it according to the following criteria: a manual inspection of spectra was carried out wherever identification was supported among 90−95% confidence values. Fold changes were calculated by comparing iTRAQ reporter ion intensities of peptides derived from AS and control valves (representative peptide MS/MS spectra and reporter ions of some differentially expressed proteins are shown in Fig. 2). Using 1.4-0.71 fold ratio as a cutoff to assign up or down-regulated proteins, we observed 56 differential proteins present in the four runs (mixtures) of the experiment, 34 up-regulated and 19 down-regulated in stenotic aortic valves, in addition to 3 proteins (haptoglobin, peptidyl-prolyl cis trans isomerase A, L-lactate dehydrogenase A chain isoform 1) which were increased or decreased depending on the isoform identified (Tables 1 and 2).
(A) Example of one of the proteins identified after iTRAQ labeling and LC-MS/MS analysis. This protein was up-regulated in aortic stenosis (labeled with 115 and 117), with respect to control valves (labeled with 114 and 116). (1) MS/MS spectra after collision-induced dissociation (CID), (2) MS/MS fragmentation spectra after high-energy collision dissociation (HCD) for the analysis of low molecular masses, (3) detail of low molecular weight area after HCD fragmentation, showing the peaks that correspond to the reporter ions. (B) Quantification of reporter ion intensities for one of the proteins varied after iTRAQ label, biglycan. As can be observed, the ratio (114 + 116)/(115 + 117), in this case corresponding to controls/AS, is lower than the 0.71 cutoff, which means this protein is increased in AS.
Annotation and functional enrichment
Functional analysis of the altered proteins was performed using DAVID v6.7. Molecular functions and biological processes were explored through the functional annotation tool to generate clusters of overrepresented Gene Ontology (GO) terms. This analysis showed 13 significant clusters where the most significant cluster was formed by proteins from the extracellular region or matrix. This cluster is composed of 24 proteins of which 13 correspond to ECM proteins. Among most significantly enriched terms we observed oxidative stress, inflammatory response and proteins related to muscle and cytoskeleton. ECM proteins together with oxidative stress proteins are primarily involved in the biological AS process (Fig. 3A).
(A) Biological implications of the 56 differentially expressed proteins according to the functional annotation clustering. (B,C) Protein-protein interaction networks were studied using the STRING v9.1 web-tool. Analyses of the 56 dysregulated proteins (B) and ECM proteins (C) are shown. Proteins with the same circle colours correspond to the same cluster, according to the classification performed using this tool.
Protein-protein interaction
The 56 deregulated proteins determined in the iTRAQ experiment were introduced into the web-tool STRING v9.1 to generate protein-protein interaction networks (Fig. 3B). We selected exclusively the 13 ECM proteins which appeared altered in the iTRAQ experiment attending to the functional clustering and we analyzed them by web-tool STRING v9.1 too. (Fig. 3C).
After the clustering of ECM proteins, we found that the biggest group was composed by 5 proteins, all of which are tightly connected to each other (Fig. 3C, red cluster). Among these differentially expressed proteins were biglycan (BGN) with a control/patient ratio of 0.70, periostin (POSTN) with a ratio of 0.55 and prolargin (PRELP) with a ratio of 0.59. These 3 proteins were identified in the four iTRAQ experiments (mixtures) and were selected for the validation of the quantitative proteomic analysis.
Validation of the quantitative proteomic analysis
SRM
To validate the results obtained by proteomic analysis, the 3 proteins with altered expression profiles were monitored by SRM in an independent group of samples (n = 10 controls, n = 10 AS patients).
For each protein, two peptides were quantified by SRM; in the case of periostin and prolargin, these peptides were found to differ significantly in their expression between groups (Fig. 4; Panel A = periostin, peptide 1: p = 0.0022 and peptide 2: p = 0.0053 and panel B = prolargin, peptide1: p = 0.0086 and peptide 2: p = 0.0481) Unfortunately, we were not able to detect the peptides that correspond to biglycan by this technique.
Validation of differential proteins by SRM analysis.
Graphical representation of the abundance (left panels) and representative extracted ion chromatograms (right panels) of the transitions observed for periostin (A) and prolargin (B). Significant increases (p < 0.05) in the expression of the measured proteotypic peptides of both proteins were observed in AS with respect to healthy valves. *p < 0.05; **p < 0.01. C: controls; AS: aortic stenosis; cps: counts per second.
Western blotting
We analyzed by WB two proteins: biglycan, not detected by SRM and periostin with the goal in mind of confirming the reliability of both techniques. Overexpression of these proteins in AS valves compared to the control valves was confirmed (p = 0.021, p = 0.025, respectively) (Fig. 5).
Discussion
Aortic stenosis is a progressive disease, often first manifesting as aortic valve sclerosis and later with valve dysfunction9. Valve dysfunction is characterized by a deregulation that depends upon the combined relationship between ECM, valve cells and valve mechanics3. In fact, alterations in the normal arrangement or composition of ECM significantly and detrimentally impact valve function producing a positive feedback loop. For this reason, there has been an increasing effort to characterize the ECM proteins in the context of AS. The study of proteins of interest by proteomic approaches is an excellent tool for the identification of tissue damage associated biomarkers, which may serve to specifically identify patients with poor prognosis10, as well as to understand the physiological/pathological processes leading to the development of AS. Following this idea we could remark that, until now, the sole comparative proteomic analyses of AS tissue valves have been recently performed by our group6,7, with the aim of understanding the molecular mechanisms of this pathology by 2-D DIGE analysis of the aortic valve tissue and by LC-MS/MS to characterize valve secretome. Along the same line as these studies, we have identified a considerable amount of differential proteins involved in ECM processes in our study by iTRAQ, which strongly supports a deep involvement of ECM remodeling in AS pathogenesis. For this reason and taking into account the complementarity of both techniques, we have supplemented our previous results with tissue secretome, with iTRAQ quantitative proteomics. Regarding this, we could broaden the knowledge about the disease by integrating the information related to secreted proteins with that of the molecular processes taking place inside the tissue.
With a cutoff of 1.4-fold upregulation and 0.71-fold downregulation, we could find 56 proteins to be significantly varied. Among these proteins, we found 13 proteins involved in ECM, with an outstanding cluster of 5 proteins formed by biglycan, periostin and prolargin, decorin and lumican. Of these proteins, 3 of them were validated by orthogonal techniques (WB and/or SRM). Periostin was selected for validation due to the controversy that exists in the literature about the role of this protein in AS11,12. On the other hand, biglycan and prolargin have been related to the atherosclerotic lesion. Nowadays, the relationship between AS and atherosclerosis remains unclear13 and therefore a deeper study of these proteins would favor to elucidate this link. Prolargin, lumican and decorin belong to the small leucine-rich proteoglycan (SLRPs) family and are closely related. Nevertheless, in contrast with lumican and decorin, the evidence about the involvement of prolargin in either atherosclerosis or AS is very limited. Besides, lumican was previously described and validated in AS patients by our group7.
If we focus on the molecular implications of these findings, we realize that periostin (POSTN) is a secreted protein expressed in cardiac fibroblasts, implicated in cardiac dysfunction14,15. It was first identified in bone, where it plays a key role regulating adhesion and differentiation of osteoblasts16,17. More recently, it has been suggested an involvement of POSTN in heart valve morphogenesis18,19,20 and cell migration12,21. Furthermore, Snider et al.20, demonstrated, that POSTN is an intriguing matrix effector protein that is required for normal ECM deposition within the heart and suggests that POSTN may be required to enhance the fibrotic remodeling effects of TGFβ and stabilize microfibril networks within the cardiac skeleton, too. Our results showing over-expression of POSTN in stenotic valves point out a critical role of this protein in AS development, where it may contribute to small vessel formation as a result of its angiogenic activity22,23. It is important to note that this protein might enhance the penetration of microvessels into the subendothelial and fibrosa layers, which are the areas where the main anatomopathological alterations take place in all types of valvular heart disease as reported by recent studies23,24. In particular, POSTN expression was detected in the subendothelial superficial layer of normal cardiac valves and it was greatly increased in the areas of neoangiogenesis24 in the degenerated valves of patients with valvular heart disease, such as those with stenotic or rheumatic valves. In addition, POSTN may contribute to valve calcification via its ability to provoke osteoblast differentiation since myofibroblasts differentiate to osteoblast-like cells in severe AS valves25.
Regarding biglycan (BGN), a small leucine-rich proteoglycan which organizes the fibril collagen assembly26, it has been defined as one of the major extracellular proteoglycans found in human atherosclerotic lesions27,28. Furthermore, it is considered an early initiator of AS lesion, contributing to wall thickening29. Previous reports have described a positive stimulation effect of pro-inflammatory factors in the ability of macrophage cells to synthesize BGN in atherosclerosis and in hypertension30. Thus, BGN levels are correlated with IL-6, TNF-α, fibrinogen and CRP plasma levels, suggesting that BGN might be involved in vascular inflammation31. It has been recently reported that macrophage cells are able to produce BGN upon stimulation by inflammatory factors26. Biglycan itself has been demonstrated to modulate the immune and inflammatory responses through the interaction with Toll-Like Receptor (TLRs) signaling cascade. Therefore, during inflammation, BGN may be released to the ECM or alternatively could be induced in macrophage cells by cytokines, to signal through the interaction with TLRs. Subsequent expression of TNFα might result in increased recruitment of macrophages that release further BGN, favoring the creation of a cycle that is capable to drive an inflammatory response in both an autocrine and a paracrine manner32. Recently, TLRs have been detected in stenotic valves33, in particular in valve interstitial cells (VICs)34, the main cellular component of the aortic valve. Moreover, studies indicate that TLRs may play a significant role in atherosclerosis35,36. Studies with animal models have demonstrated that BGN binds to murine TLR 2 and 4, whereby it induces the production of inflammatory cytokines through the nuclear factor kappa B (NF-kB) pathway30. Taking into account the already postulated relationship between atherosclerosis and AS37 based on histological findings that point to tissue mineralization, accumulation of lipids and inflammation38, the observed up-regulation of BGN in AS valves found in this work is in concordance with previous studies and suggests this proteoglycan as an early initiator of the sclerotic lesion through an interaction with TLRs within the aortic valve.
In the same way, prolargin protein (PRELP) was found up-regulated in AS valve tissue39,40. This protein is present in all types of connective tissue. As the rest of the SLRPs, PRELP has a great affinity for the collagens I and II through its leucine-rich repeat domain41, joins fibroblasts through its N-terminal and plays an important role in bone remodeling control42. Thus, PRELP could exert a similar effect to BGN in the development of AS calcification, together with other proteoglycans such as lumican and decorin43,44, which have been also found increased in AS in the iTRAQ analysis. The increase in AS of all the SLRPs detected in the present proteomic study may serve to fundament, once again, the relationship of this pathology with atherosclerosis36, given that lumican, decorin and prolargin are proteins involved in both pathologies which show similar expression trends43.
The main limitation of our study is the low number of samples included. Despite the fact that validation was performed in an independent cohort of subjects and by two different orthogonal techniques, it would be recommendable to increase the number of patients and controls in order to confirm the potential value of the hereby described protein indicators. Moreover, a prospective study will be required to determinate the clinical utility of these proteins as potential biomarkers which could allow distinguishing patients at risk of developing degenerative AS. In conclusion, our results state the importance of the extracellular matrix remodeling in AS. The use of a comprehensive analysis of the aortic valve proteome using 2D-LC MS/MS iTRAQ methodology has allowed the identification of a specific protein profile linked to the disease, highlighting the importance of the extracellular matrix proteins. Specifically, biglycan, periostin and prolargin have been pointed out as proteins of interest and have been validated using orthogonal techniques (WB and/or SRM). These results support the existence of a dynamic extracellular matrix remodeling in human aortic valves during AS development.
Material and Methods
Patient and control subjects’ selection
In this work, we employed the same samples used in our previous secretome study6. We obtained as many subjects as possible for this kind of proteomics analysis and we eliminated any potential bias inherent in their selection which meant a lot of effort over time, mainly in control samples. Heart valves with degenerative AS (n = 18) were obtained from patients of both sexes (50% male, 50% female), with an average age of 74 ± 4 years, who underwent aortic valve replacement due to severe degenerative AS. All patients (100%) had hypertension, whereas 55% suffered hyperlipemia and 66% diabetes mellitus. Coronary artery disease was present in 55% patients and surgery was indicated according to current clinical practice guidelines. Patients with aortic regurgitation, mitral valve disease or any suspicion of rheumatic disease were not included in the study. Control valves were obtained from necropsies (n = 18). Before resecting the valve, a cardiac surgeon explored it carefully. Valves with calcifications or cusp restriction were excluded as control cases. If inserted saline in the aortic root remained in the closed valve, we ruled out significant regurgitation and we took these aortic valves as control cases. These subjects did not die from cardiovascular illnesses and had no history of coronary artery disease or diabetes mellitus (Table 3).
The study was conducted according to the recommendations of the Declaration of Helsinki and was approved by the ethics committee of Hospital Virgen de la Salud (Toledo, Spain). Signed informed consent was obtained from all subjects or relatives in case of necropsies prior to their inclusion in the study.
Tissue Sample preparation
Aortic valves were processed according to Martin-Rojas et al.6 with minor modifications.
Briefly, AS valves were processed within a maximum of 2 hours after surgery having maintained the tissue at 4 °C in RPMI medium. The valves were washed 3 times in PBS to reduce blood contaminants. One aortic valve leaflet was then ground into powder in liquid N2 with a mortar and 0.2 g of this powder were resuspended in 400 μl of protein extraction buffer (10 mM Tris [pH 7.5], 500 mM NaCl, 0.1% Triton x-100, 1% β-mercaptoethanol, 1 mM PMSF)45,46. The homogenate was centrifuged at 21,000 g (5840R Eppendorf) for 15 min at 4 °C to precipitate the membranes and tissue debris and the supernatant (E1) containing most of the soluble proteins, was collected and stored at −20 °C. The pellet was then solubilized in 7M Urea, 2M Thiourea, 4% CHAPS47,48 and centrifuged again at 21,000 g. This second supernatant (E2) was collected and the cellular debris and lipids were eliminated with the pellet. Both protein extracts (E1 and E2) were pooled and the protein concentration was determined by the Bradford-Lowry method (Bio-Rad protein assay)49.
iTRAQ labeling and SCX fractionation
iTRAQ labeling was carried out with an iTRAQ reagent 4plex Protein Quantitation kit (AB Sciex) according to manufacturer’s protocol. The amount of 100 μg of each aortic valve lysate was dissolved in 0.5 M triethylammonium bicarbonate (TEAB) (pH 8.5). Aortic valve proteins were reduced with 2 μL of (tris(2-carboxyethyl) phosphine) (TCEP) at 60 °C for 1 h and alkylated with 1 μL methyl methanethiosulfonate (MMTS) for 10 min at room temperature. Samples were digested using trypsin (porcine Promega) (1:40, w/w) for 16 h at 37 °C). Digested samples were further processed by labeling the derived peptides from control aortic valves with 114 and 116 iTRAQ labels and from AS valves with 115 and 117, thus providing technical replicates within a single run (Table SM2).
The iTRAQ labeled peptides were pooled and fractionated by 2D-LC with strong cation exchange chromatography (SCX) followed by a C-18 reversed phase (RP) nano-column (100 μm I.D. and 12 cm, Mediterranea sea, Teknokroma) and analyzed in a continuous acetonitrile gradient consisting of 0–43% B in 140 min, 50−90% B in 1 min (B = 95% acetonitrile, 0.5% acetic acid). A flow rate of 300 nL/min was used to elute peptides from the RP nano-column to an emitter nanospray needle for real time ionization and peptide fragmentation on an LTQ Orbitrap XL ETDTM mass spectrometer (Thermo Fisher). An enhanced FT-resolution spectrum (resolution = 60000) followed by the MS/MS spectra from most intense five parent ions were analyzed along the chromatographic run (180 min). Dynamic exclusion was set at 0.5 min. Expression changes were calculated using the reporter ion intensities (peaks areas) as control (114 + 116)/AS patients (115 + 117).
Protein identification and quantification were performed with the Protein Discoverer 1.0 software (Thermo Fisher) using MASCOT as the search engine. Two mixed cleavages were allowed and an error of 15 ppm or 0.8 Da was set for full MS or MS/MS spectra searches, respectively. Decoy database search for FDR analysis was set at 0.05 by applying corresponding filters. Phosphorylation in Serine, Threonine, or Tyrosine was specified as variable modification due to the importance that this mechanism has in matrix remodeling50.
Bioinformatics analysis of identified proteins
For a functional examination of the identified proteins, the list of the 56 significantly varied proteins was implemented on the on-line software David Bioinformatics Resources 6.7 (NIH)51 and Search Tool for the Retrieval of Interacting Genes/Proteins (STRING v9.1), for functional and protein interaction analyses, respectively.
Functional annotation clustering was performed in order to avoid redundancy of enriched categories and pathways (data not shown).
Selected Reaction Monitoring (SRM)
Protein samples (10 AS patients and 10 controls) were reduced with 100 mM dithiothreitol (DTT, Sigma-Aldrich) in 50 mM ammonium bicarbonate (99% purity; Scharlau) for 30 min at 37 °C and alkylated for 20 min at room temperature (RT) with 550 mM iodoacetamide (Sigma-Aldrich) in 50 mM ammonium bicarbonate. The proteins were then digested in 50 mM ammonium bicarbonate, 15% acetonitrile (LC-MS grade, Scharlau) with sequencing grade modified porcine trypsin at a final concentration of 1:50 (trypsin:protein). After overnight digestion at 37 °C, 2% formic acid (FA, 99.5% purity, Sigma-Aldrich) was added and the samples were cleaned with Pep-Clean spin columns (Pierce) according to the manufacturer’s instructions. Tryptic digests were dried in speed-vac and resuspended in 2% acetonitrile, 2% FA prior to MS analysis. The LC-MS/MS system consisted of a TEMPO nano LC system (Applied Biosystems) combined with a nano LC autosampler coupled to a modified triple quadrupole (4000QTRAP LC/MS/MS, Applied Biosystems). Three replicate injections (2 μg of protein in 4 μL) were performed per sample in the mobile phase A (2% ACN/98% water, 0.1% FA) at a flow rate of 10 μL/min for 5 min. Peptides were loaded onto a μ-Precolumn Cartridge (Acclaim Pep Map 100 C18, 5 μm, 100 Å; 300 μm i.d. X 5 mm, LC Packings) to preconcentrate and desalt samples. Reversed-phase liquid chromatography (RPLC) was performed on a C18 column (Onyx Monolithic C18, 150 × 0.1 mm I.D., Phenomenex) in a gradient of phase A and phase B (98% ACN/2% water, 0.1% FA). The peptides were eluted at a flow rate of 300 nL/min in a continuous acetonitrile gradient: 2–15% B for 2 min, 15–50% B for 38 min, 50 to 90% B for 2 min and 90% B for 3 min. Both the TEMPO nano LC and 4000QTRAP system were controlled by Analyst Software v.1.4.2. The mass spectrometer was set to operate in positive ion mode with ion spray voltage of 2800 V and a nanoflow interface heater temperature of 150 °C. The source and curtain gas were set to 20 and 10 psi respectively and nitrogen was applied as both curtain and collision gas.
Theoretical SRM transitions were designed using MRMpilot software v1.1 (ABSciex), with the following settings: Enzyme = trypsin, missed cleavages = 0; modifications in peptide ≤ 3; charge states = +1 from 300 to 600 Da, +2 from 500 to 2000 Da, +3 from 900 to 3000 Da, +4 from 1600 to 4000 Da, +5 from 2400 to 10000 Da; studied modification = none; fixed modifications = carboxyamidomethylation; variable modifications = none; min. number of amino acids ≥ 5; max. number of amino acids ≤30; ignore multiple modification sites; 3 transitions per peptide (Table 4).
A pool containing a mixture of all the samples was digested as described previously and analyzed in the 4000QTrap using a MIDAS acquisition method that included the theoretical transitions. Transitions were selected when the three co-eluting peaks (corresponding to the three transitions of the same peptide) had a signal-to-noise ratio over 5 and the MS/MS data matched the theoretical spectrum for that peptide. Collision energy was optimized to obtain the maximum transmission efficiency and sensitivity for each SRM transition. A total of 12 transitions (3 per peptide) were monitored during an individual sample analysis. They were acquired at unit resolution in both Q1 and Q3, with dwell times of 50 ms resulting in cycle times of 1.8 s. The IntelliQuan algorithm included in Analyst 1.4.2 software was used to calculate the peptide abundance on the basis of peak areas after integration.
Western blotting (WB)
Protein samples obtained from 4 AS and 4 control valves were resolved by 12% SDS-PAGE using a Miniprotean II electrophoresis unit (Bio-Rad) run at a constant current of 25 mA/gel during 1 hour. After SDS-PAGE, the proteins were transferred to a nitrocellulose membrane under a constant voltage of 15 V for 20 minutes. To ensure that equal amounts of aortic valve tissue were loaded in every polyacrylamide gel well (1-DE Western blotting) Ponceau S staining was performed on the transferred membranes. The membranes were then blocked with 7.5% non-fat milk for 1 hour6. Next, they were incubated overnight at 4 °C with the primary antibody followed by a 1 hour RT incubation with the specific HRP-conjugated secondary antibody, both in PBS-T containing 5% of non-fat dry milk. Detection was performed by enhanced chemiluminescence (ECL, GE Healthcare) following the manufacturers’ instructions.
The primary antibodies for Western blots included rabbit polyclonal antisera against periostin (catalog number: ab14041) and a monoclonal antibody against biglycan (catalog number: ab54855), all from Abcam (Abcam).
Statistical analysis
Western Blot bands were measured using a GS-800 Calibrated Densitometer (Bio-Rad) and the values obtained analyzed using the statistical software packages SPSS 13.0 and GraphPad InStat (GraphPad software). A Kolmogorov-Smirnov test was applied to evaluate normal distribution of the population analyzed. When normal distribution was demonstrated, a Levene test for homogeneity of variance was performed and the Student t-test was used to compare band intensities and SRM peak areas, selecting adequate p-value in view of homogeneity results. For populations with no normal distribution, the non-parametric Mann-Whitney U test was used. For all tests, statistical significance was accepted when p < 0.05. All LC-MS/MS identifications were performed by Decoy database search for FDR analysis set at 0.05 by applying corresponding filters (Mascot Significance Threshold and Score versus Charge state).
Disclosures
None
Additional Information
How to cite this article: Martin-Rojas, T. et al. iTRAQ proteomic analysis of extracellular matrix remodeling in aortic valve disease. Sci. Rep. 5, 17290; doi: 10.1038/srep17290 (2015).
References
Maganti, K., Rigolin, V. H., Sarano, M. E. & Bonow, R. O. Valvular heart disease: Diagnosis and management. Mayo Clin Proc. 85, 483–500 (2010).
Cowell, S. J., Newby, D. E., Boon, N. A. & Elder, A. T. Calcific aortic stenosis: same old story? Age Ageing. 33(6), 538–44 (2004).
Wiltz, D. et al. Extracellular matrix organization, structure and function in Calcific aortic valve disease (Ed. Aikawa, E. ), 3–30 (In Tech, 2013).
Hinton, R. B. et al. Extracellular matrix remodeling and organization in developing and diseased aortic valve. Circ Res. 98(11), 1431–8 (2006).
Kaden, J. J. et al. Inflammatory regulation of extracellular matrix remodeling in calcific aortic valve stenosis. Cardiovasc Pathol. 14(2), 80–7 (2005).
Alvarez-Llamas, G. et al. Modification of the secretion pattern of proteases, inflammatory mediators and extracellular matrix proteins by human aortic valve is key in severe aortic stenosis. Mol Cell Proteomics. 12(9), 2426–39 (2013).
Martín-Rojas, T. et al. Proteomic profile of human aortic stenosis: insights into the degenerative process. J Proteome Res. 11(3), 1537–50 (2012).
Wu, W. W., Wang, G., Baek, S. J. & Shen, R. F. Comparative study of three proteomic quantitative methods, DIGE, cICAT and iTRAQ, using 2D gel- or LC-MALDI TOF/T1OF. J Proteome Res. 5(3), 651–8 (2006).
Otto, C. M. Valvular aortic stenosis: disease severity and timing of intervention. J Am Coll Cardiol. 47, 2141–2151 (2006).
Cubedo, J., Padró, T. & Badimon, L. Coordinated proteomic signature changes in immune response and complement proteins in acute myocardial infarction: the implication of serum amyloid P-component. Int J Cardiol. 168, 5196–5204 (2013).
Tkatchenko, T. V. et al. Lack of periostin leads to suppression of Notch1 signaling and calcific aortic valve disease. Physiol Genomics. 39(3), 160–8 (2009).
Hakuno, D. et al. Periostin advances atherosclerotic and rheumatic cardiac valve degeneration by inducing angiogenesis and MMP production in humans and rodents. J Clin Invest. 120(7), 2292–306 (2010).
Novaro, G. & Griffin, B. Calcific aortic stenosis: another face of atherosclerosis? Cleve Clin J Med. 70, 1–7 (2013).
Katsuragi, N. et al. Periostin as a novel factor responsible for ventricular dilation. Circulation. 110(13), 1806–13 (2004).
Litvin, J. et al. Periostin and periostin-like factor in the human heart: possible therapeutic targets. Cardiovasc Pathol. 15, 24–32 (2006).
Takeshita, S., Kikuno, R., Tezuka, K. & Amann, E. Osteoblast-specific factor 2: cloning of a putative bone adhesion protein with homology with the insect protein fasciclin I. Biochem J. 294, 271–8 (1993).
Horiuchi, K. et al. Identification and characterization of a novel protein, periostin, with restricted expression to periosteum and periodontal ligament and increased expression by transforming growth factor beta. J Bone Miner Res. 14(7), 1239–49 (1999).
Norris, R. A. et al. Identification and detection of the periostin gene in cardiac development. Anat Rec A Discov Mol Cell Evol Biol. 281(2), 1227–33 (2004).
Kruzynska-Frejtag, A., Machnicki, M., Rogers, R., Markwald, R. R. & Conway, S. J. Periostin (an osteoblast-specific factor) is expressed within the embryonic mouse heart during valve formation. Mech Dev. 103(1–2), 183–8 (2001).
Snider, P. et al. Periostin is required for maturation and extracellular matrix stabilization of noncardiomyocyte lineages of the heart. Circ Res. Apr 11, 102(7), 752–60 (2008).
Roy, S. et al. Transcriptome-wide analysis of blood vessels laser captured from human skin and chronic wound-edge tissue. Proc Natl Acad Sci USA 104(36), 14472–14477 (2007).
Shao, R. et al. Acquired expression of periostin by human breast cancers promotes tumor angiogenesis through up-regulation of vascular endothelial growth factor receptor 2 expression. Mol Cell Biol. 24(9), 3992–4003 (2004).
Siriwardena, B. S. et al. Periostin is frequently overexpressed and enhances invasion and angiogenesis in oral cancer. Br J Cancer. 95(10), 1396–1403 (2006).
Lorts, A., Schwanekamp, J. A., Elrod, J. W., Sargent, M. A. & Molkentin, J. D. Genetic manipulation of periostinexpression in the heart does not affect myocytecontent, cell cycle activity, or cardiac repair. Circ Res. 104(1), e1–e7 (2009).
Rajamannan, N. M. et al. Human aortic valve calcification is associated with an osteoblast phenotype. Circulation. 107, 2181–2184 (2003).
Ameye, L. et al. Abnormal collagen fibrils in tendons of biglycan/fibromodulin-deficient mice lead to gait impairment, ectopic ossification and osteoarthritis. FASEB J. 16, 673–80 (2002).
O’Brien, K. D. et al. Comparison of apolipoprotein and proteoglycan deposits in human coronary atherosclerotic plaques: co-localization of biglycan with apolipoproteins. Circulation. 98, 519–527 (1998).
Olin, K. L., Potter-Perigo, S., Barrett, P. H., Wight, T. N. & Chait, A. Biglycan, a vascular proteoglycan, binds differently to HDL2 and HDL3: role of ApoE. Arterioscler Thromb Vasc Biol. 21, 129–135 (2001).
Nakashima, Y., Wight, T. N. & Sueishi, K. Early atherosclerosis in humans: role of diffuse intimal thickening and extracellular matrix proteoglycans. Cardiovasc Res. 79, 14–23 (2008).
Alimohamad, H., Habijanac, T., Larjava, H. & Häkkinen, L. Colocalization of the collagenbinding proteoglycans decorin, biglycan, fibromodulin and lumican with different cells in human gingiva. J Periodontal Res. 40, 73–86 (2005).
Sardo, M. A. et al. Biglycan expression in hypertensive subjects with normal or increased carotid intima-media wall thickness. Clin Chim Acta. 406, 89–93 (2009).
Schaefer, L. et al. The matrix component biglycan is proinflammatory and signals through Toll-like receptors 4 and 2 in macrophages. J Clin Invest. 115, 2223–2233 (2005).
Meng, X. et al. Expression of functional Toll-like receptors 2 and 4 in human aortic valve interstitial cells: potential roles in aortic valve inflammation and stenosis. Am J Physiol Cell Physiol. 294, C29–C35 (2008).
Babu, A. N. et al. Lipopolysaccharide stimulation of human aortic valve interstitial cells activates inflammation and osteogenesis. Ann Thorac Surg. 86, 71–76 (2008).
Xu, X. H. et al. Toll-like receptor-4 is expressed by macrophages in murine and human lipid-rich atherosclerotic plaques and upregulated by oxidized LDL. Circulation. 104, 3103–3108 (2001).
Derbali, H. et al. Increased biglycan in aortic valve stenosis leads to the overexpression of phospholipid transfer protein via Toll-like receptor 2. Am J Pathol. 176(6), 2638–45 (2010).
Rajamannan, N. M. Calcific aortic stenosis: lessons learned from experimental and clinical studies. Arterioscler Thromb Vasc Biol. 29, 162–168 (2009).
Mohty, D. et al. Association between plasma LDL particle size, valvular accumulation of oxidized LDL and inflammation in patients with aortic stenosis. Arterioscler Thromb Vasc Biol. 28, 187–193 (2008).
Kobe, B. & Deisenhofer, J. The leucine-rich repeat, a versatile binding motif. Trends Biochem Sci. 19, 415–421 (1994).
Grover, J. & Roughley, P. J. Characterization of the human proline/arginine-rich end leucine-rich repeat protein (PRELP) gene promoter and identification of a repressor element. Biochem J. 336, 77–82 (1998).
Bengtsson, E. et al. The leucine-rich repeat protein PRELP binds perlecan and collagens and may function as a basement membrane anchor. J Biol Chem. 277, 15061–15068 (2002).
Rucci, N. et al. The glycosaminoglycan-binding domain of PRELP acts as a cell type–specific NF-kb inhibitor that impairs osteoclastogenesis. J Cell Biol. 187, 669–683 (2009).
Gutierrez, P. et al. Differences in the distribution of versican, decorin and biglycan in atherosclerotic human coronary arteries. Cardiovasc Pathol. 6, 271–278 (1997).
Schaefer, L. & Schaefer, R. M. Proteoglycans: from structural compounds to signaling molecules. Cell Tissue Res. 339, 237–246 (2010).
Gil-Dones, F. et al. Development of an optimal protocol for the proteomic analysis of stenotic and healthy aortic valves. Rev Esp Cardiol. 63, 46–53 (2010).
Barderas, M. G. et al. Serodiagnosis of African swine fever using the recombinant protein p30 expressed in insect larvae. J Virol Methods. 89, 129–136 (2000).
Barderas, M. G. et al. Circulating human monocytes in the acute coronary syndrome express a characteristic proteomic profile. J Proteome Res. 6, 876–886 (2007).
Vivanco, F. et al. Quest for novel cardiovascular biomarkers by proteomic analysis. J Proteome Res. 4, 1181–91 (2005).
Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 72, 248–254 (1976).
Sariahmetoglu, M. et al. Regulation of matrix metalloproteinase-2 (MMP-2) activity by phosphorylation. FASEB J. 21(10), 2486–95 (2007).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Acknowledgements
We thank the CNIC (Centro Nacional Investigaciones Cardiovasculares) for assistance with the protein identification. This work was supported by grants from the Instituto de Salud Carlos III (FIS PI070537, PI11-02239, PI14/01917) and Redes Temáticas de Investigación Cooperativa (FONDOS FEDER, RD06/0014/1015, RD12/0042/0071). These results are lined up with the Spanish initiative on the Human Proteome Project (SpHPP).
Author information
Authors and Affiliations
Contributions
T.M. and L.M. performed the iTRAQ and the SRM analysis as well as the statistical analysis obtained with this technology. S.A. performed the validation by western-blot and the statistical analysis. E.C. and J.A.L. performed the analysis of the iTRAQ results obtained and also performed the statistical analysis. All the work related to the selection, study and follow-up of patients was carried out by L.F.L. in collaboration with L.R.P. Selection and collection of control samples was performed by E.R.L. and M.R. Finally, F.C. and M.G.B. designed and supervised the complete study.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Martin-Rojas, T., Mourino-Alvarez, L., Alonso-Orgaz, S. et al. iTRAQ proteomic analysis of extracellular matrix remodeling in aortic valve disease. Sci Rep 5, 17290 (2015). https://doi.org/10.1038/srep17290
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep17290
- Springer Nature Limited
This article is cited by
-
The contribution of amyloid deposition in the aortic valve to calcification and aortic stenosis
Nature Reviews Cardiology (2023)
-
Differential proteome profile, biological pathways, and network relationships of osteogenic proteins in calcified human aortic valves
Heart and Vessels (2022)
-
Lipid mass spectrometry imaging and proteomic analysis of severe aortic stenosis
Journal of Molecular Histology (2020)
-
Heat shock protein 90 is downregulated in calcific aortic valve disease
BMC Cardiovascular Disorders (2019)
-
Periostin in cardiovascular disease and development: a tale of two distinct roles
Basic Research in Cardiology (2018)