Abstract
The nucleolus, whose primary function is ribosome biogenesis, plays an essential role in p53 activation. Ribosome biogenesis is inhibited in response to cellular stress and several nucleolar proteins translocate from the nucleolus to the nucleoplasm, where they activate p53. In this study, we analysed precisely how impaired ribosome biogenesis regulates the activation of p53 by depleting nucleolar factors involved in rRNA transcription or rRNA processing. Nucleolar RNA content decreased when rRNA transcription was inhibited. In parallel with the reduced levels of nucleolar RNA content, the nucleolar protein Myb-binding protein 1 A (MYBBP1A) translocated to the nucleoplasm and increased p53 acetylation. The acetylated p53 enhanced p21 and BAX expression and induced apoptosis. In contrast, when rRNA processing was inhibited, MYBBP1A remained in the nucleolus and nonacetylated p53 accumulated, causing cell cycle arrest at the G1 phase by inducing p21 but not BAX. We propose that the nucleolus functions as a stress sensor to modulate p53 protein levels and its acetylation status, determining cell fate between cell cycle arrest and apoptosis by regulating MYBBP1A translocation.
Similar content being viewed by others
Introduction
Mutations in the tumour suppressor p53 gene occur in approximately 50% of all human cancers and play a pivotal role in tumour progression. p53 is maintained at low levels in unstressed cells by its interaction with E3 ubiquitin ligases such as HDM21,2,3. However, p53 is activated in response to cell stress, producing a number of downstream effects1,4,5,6,7,8. p53 induces cell cycle arrest by activating target genes such as the CDK inhibitor p21CDKN1A9,10 and induces apoptosis by activating distinct classes of target genes, including BBC3 (PUMA)11, BAX12, PMAIP1 (NOXA)13, P53AIP114 and TP53I3 (PIG3)15.
Several studies have reported the mechanisms that select p53 target genes and determine cell fate. First, the strength of p53 binding to cell cycle gene responsive elements (REs) is generally more robust compared with that of apoptosis-related genes16. Second, post-translational modifications, such as acetylation of C-terminal lysine residues17,18, acetylation of the DNA binding domain at Lys-12019,20 and phosphorylation at Ser-4614,21,22, regulate sequence-specific DNA binding of p53. Third, several p53 binding partners alter the ability of p53 to recognise specific REs and recruit transcriptional coactivators to certain loci23,24,25. Fourth, protein levels of p53 affect the expression of its target genes26. Finally, epigenetic histone modifications of p53 target promoters can influence the selection of p53 target genes27. These results demonstrate the mechanisms of how p53 produces cellular outcomes under specific stressors. However, the upstream regulators that coordinate the activation of p53 and integrate the stress response into individual outcomes have not been completely elucidated.
Recent studies have revealed that the nucleolus plays an important role in activating p5328,29. The nucleolus is a distinct subnuclear component that was originally considered the site of ribosome biogenesis, as it functions in ribosomal RNA (rRNA) transcription, rRNA processing and ribosome assembly with many ribosomal proteins. In response to cellular stress, several nucleolar proteins, including p14/p19Arf, nucleophosmin, nucleolin, nucleostemin, RPL11, RPL5, RPL23 and RPS7, translocate from the nucleolus to the nucleoplasm, where they activate p53 by inhibiting HDM230,31,32,33,34,35,36,37,38,39,40,41,42. In particular, RPL11 and RPL5, which are components of the 5 S ribonucleoprotein particle (RNP) complex, play an important role activating p53 under conditions in which ribosomal biogenesis is inhibited43,44,45. We recently reported that 5 S RNP activates p53 and induces cellular senescence in response to aberrant ribosome biogenesis under oncogenic and replicative stresses46.
In addition, we have shown that nucleolar protein Myb-binding protein 1A (MYBBP1A) acts as a nucleolar stress response molecule by regulating p53 acetylation47. Nucleolar localisation of MYBBP1A depends on nucleolar RNA content. Inhibited rRNA transcription following DNA damage or glucose starvation leads to reduced nucleolar RNA content and translocation of MYBBP1A from the nucleolus to the nucleoplasm, where it binds nonacetylated p53, facilitates the interaction between p53 and histone acetyltransferase p300 and induces p53 acetylation and tetramerisation47,48,49,50. Then, MYBBP1A dissociates from the acetylated form of p53.
Here we show that enhanced p53 acetylation via MYBBP1A correlated with the induction of apoptosis. MYBBP1A translocated from the nucleolus to the nucleoplasm when rRNA transcription was inhibited, in parallel with a reduction in nucleolar RNA content and elevated p53 acetylation. Acetylated p53 induced apoptosis, accompanied by high p21 and BAX expression levels. In contrast, nucleolar RNA content increased and MYBBP1A remained in the nucleolus when rRNA processing was inhibited. In this case, a nonacetylated form of p53 accumulated via RPL11, which caused cell cycle arrest at the G1 phase by inducing p21. These results suggest that the reduction in nucleolar RNA content strictly regulates MYBBP1A translocation to the nucleoplasm, thereby determining cell fate between cell cycle arrest and apoptosis.
Results
Reduced levels of rRNA transcription correlate with the induction of apoptosis
Impaired ribosome biogenesis in response to cellular stress reportedly leads to the activation of p5328,51. Activated p53 determines cell fate (e.g. cell cycle arrest or apoptosis) by inducing distinct classes of target genes1. However, the relationship between impaired ribosome biogenesis and the p53-mediated cell fate decision remains to be elucidated.
The proteins implicated in ribosome biogenesis were depleted to investigate the relationship between impaired ribosome biogenesis and p53 activation status. First, we depleted several rRNA transcription factors, such as polymerase RNA 1 polypeptide B (POLR1B), upstream binding factor (UBF), polymerase RNA 1 polypeptide A (POLR1A) and transcription initiation factor IA (TIF-IA) in MCF-7 cells (Fig. 1a,b). Downregulating these factors inhibited rRNA transcription, as shown by the reverse transcription-quantitative polymerase chain reaction (Fig. 1c) and Northern blotting analyses using an internal transcribed sequence 1 probe (Fig. 1d). Similar results were obtained in U2OS cells (Fig. S2a and S2b). The reduced rate of rRNA transcription differed among the factors that were depleted (Figs. 1c,d, S2a and S2b). The total amount and K382-acetylation of p53 increased and were correlated with a reduced rRNA transcription rate (Figs. 1c–e and S2a–c). p21 and BAX are also increased in parallel with the activation of p53 and inhibition of rRNA transcription (Figs. 1c–e and S2a–c). Elevated p21 and BAX levels were abrogated by depleting p53 (Figs. 1e and S2c).
Gradual reduction in rRNA transcription correlates with the activation of p53 and the induction of apoptosis.
(a–d) MCF-7 cells were transfected with siCtrl, siPOLR1B, siUBF, siPOLR1A or siTIF-IA. (a) Expression of rRNA transcription factors was assessed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) at 48 h after transfection (n = 3). Error bars indicate mean ± standard deviation (SD). (b) Protein levels of rRNA transcription factors were assessed by immunoblotting at 48 h after transfection. (c) pre-rRNA transcription was assessed by RT-qPCR at 60 h after transfection (n = 3). Error bars indicate mean ± SD. (d) Northern blotting was performed at 60 h after transfection using a probe specific for the rRNA internal transcribed spacer 1 region. An ethidium bromide-stained gel is shown at the bottom as a loading control. (e–g) MCF-7 cells were transfected with combinations of siRNAs as indicated. (e) Cell lysates were prepared at 60 h after transfection and immunoblotted using the indicated antibodies. Lysates from 2 μg/ml camptothecin (CPT)-treated cells were used as a positive control. Asterisk indicates nonspecific bands. (f) DNA content was determined by flow cytometry at 72 h after transfection (n = 3). Error bars indicate mean ± SD. (g) Cell lysates were prepared at 72 h after transfection and immunoblotted using the indicated antibodies. Lysates from 2 μg/ml CPT-treated cells were used as a positive control.
Next, we tested the effect of depleting the rRNA transcription factors on cell cycle arrest at the G1 phase or apoptosis because depleting these factors upregulated p21 and BAX protein levels. We showed that depleting these factors using fluorescence-activated cell sorting (FACS) analysis increased the ratios of cells with subG1 DNA content, which was reversed by depleting p53 (Figs. 1f, S1 and S2d). We also found the enhanced poly (ADP-ribose) polymerase (PARP) cleavage, an indicator of apoptosis (Figs. 1g and S2e). Notably, the subG1 apoptotic cell ratios and PARP cleavage were correlated with the reduced rate of rRNA transcription. The cells with G1 DNA content were increased in MCF-7 cells (Fig. 1f) but not in U2OS cells (Fig. S2d). Taken together, these results suggest that the total amount and acetylation of p53 increased and that activated p53 caused apoptosis in response to a decrease in rRNA transcription.
Gradual reduction in nucleolar RNA content leads to translocation of MYBBP1A, p53 acetylation and apoptosis in a dose-dependent manner
We reported previously that inhibiting rRNA transcription decreases nucleolar RNA content and causes translocation of the nucleolar protein MYBBP1A to the nucleoplasm, where it enhances K382-acetylation of p53 by stabilising the association between p53 and p30047,48.
In this study, we evaluated whether MYBBP1A enhances p53 acetylation, which was negatively correlated with rRNA transcription levels. We first confirmed that nucleolar RNA content decreased in parallel with the reduction in rRNA transcription when several rRNA transcription factors were depleted in MCF-7 cells (Fig. 2a). Next, we examined subcellular localisation of MYBBP1A. MYBBP1A was localised predominantly in the nucleolus of control cells, whereas it translocated from the nucleolus to the nucleoplasm in response to the depletion of rRNA transcription factors (Fig. 2b). Notably, more MYBBP1A translocated from the nucleolus with lower nucleolar RNA content (Fig. 2a,b).
p53 acetylation was enhanced by the nucleolar protein Myb-binding protein 1A (MYBBP1A) in parallel with a reduction in nucleolar RNA content.
(a) MCF-7 cells were transfected with siCtrl, siPOLR1B, siUBF, siPOLR1A or siTIF-IA. Nucleolar RNA content was spectrophotometrically quantified at 60 h after transfection (n = 3). Nucleolar RNA content of control cells was normalised to 100%. Error bars indicate mean ± standard deviation. (b) MCF-7 cells were transfected with the indicated siRNAs. Immunofluorescence staining was performed using the indicated antibodies at 60 h after transfection. Upstream binding factor (UBF) was used as a nucleolar marker. (c) MCF-7 cells were transfected with combinations of siRNAs as indicated for 60 h. Cell lysates were prepared from these cells and were immunoblotted using the indicated antibodies. Lysates from 2 μg/ml camptothecin-treated cells were used as the positive control.
Then, we tested whether MYBBP1A is required for the gradual activation of p53 when several rRNA transcription factors were depleted, because translocated MYBBP1A activates p5347,48. As expected, the activation of p53 and the accumulation of p21 and BAX were counteracted by depleting MYBBP1A (Fig. 2c). Depleting RPL11, a component of the 5 S RNP complex, also reversed these phenomena (Fig. 2c). This observation is consistent with reports that 5 S RNP components induce the accumulation of p53 by inhibiting HDM2 and that they are required to translocate MYBBP1A to the nucleoplasm47,48. Similar results were observed in U2OS cells (Fig. S3). Thus, it is likely that p53 is activated via MYBBP1A and RPL11 in response to impaired rRNA transcription.
We showed that p53 acetylation and the induction of apoptosis correlated with a decrease in nucleolar RNA content when several rRNA transcription factors were depleted (Figs. 1,2, S2 and S3). However, we cannot exclude the possibility that other signals or pathways are involved in the activation of p53 and the induction of apoptosis, besides the reduction in nucleolar RNA content. Therefore, we treated MCF-7 cells with various concentrations of siTIF-IA and tested the relationship between reduced nucleolar RNA content and the activation of p53. TIF-IA levels were negatively correlated with siTIF-IA concentration (Fig. 3a,b). Pre-rRNA levels and nucleolar RNA content also decreased in an siTIF-IA concentration-dependent manner (Fig. 3c,d). MYBBP1A translocated from the nucleolus to the nucleoplasm in proportion to the reduction in nucleolar RNA content (Fig. 3e). The total amounts and acetylation status of p53, p21 and BAX (Fig. 3f), cells in the subG1 phase (Figs. 3g and S4) and PARP cleavage (Fig. 3h) increased in line with the reduced nucleolar RNA content. These phenomena (i.e. the enhancement of total amounts and acetylation of p53, p21, BAX, cells in the subG1 phase and PARP cleavage) were reversed in the presence of siMYBBP1A (Figs. 3f–h and S4). Similar results were observed when MCF-7 cells were treated with another siRNA for TIF-IA (Fig. S5) and U2OS cells were treated with siTIF-IA (Fig. S6). The cells with G1 DNA content were increased by siTIF-IA treatment in MCF-7 cells (Figs. 3g and S5e) but not in U2OS cells (Fig. S6e). Taken together, our results show that MYBBP1A, which translocated to the nucleoplasm in parallel with reduced nucleolar RNA content, activated p53 by enhancing acetylation and induced apoptosis when rRNA transcription was inhibited.
siRNA for transcriptional intermediary factor 1A (TIF-IA) treatment causes Myb-binding protein 1A (MYBBP1A)-dependent p53 acetylation and apoptosis in a dose-dependent manner.
(a–e) MCF-7 cells were transfected with the indicated concentrations of siTIF-IA. (a) TIF-IA mRNA levels were assessed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) at 60 h after transfection (n = 3). Error bars indicate mean ± standard deviation (SD). (b) Protein levels of TIF-IA were assessed by immunoblotting at 36 h after transfection. The middle panel is long exposure of the top panel. (c) pre-rRNA transcription was assessed by RT-qPCR at 60 h after transfection (n = 3). Error bars indicate mean ± SD. (d) Nucleolar RNA was isolated from purified nucleoli and quantified by spectrophotometry at 60 h after transfection (n = 3). Nucleolar RNA content of control cells was normalised to 100%. Error bars indicate mean ± SD. (e) Immunofluorescence staining was performed using the indicated antibodies at 60 h after transfection. (f–h) MCF-7 cells were transfected with combinations of siRNAs as indicated. (f) Cell lysates were prepared at 60 h after transfection and were immunoblotted using the indicated antibodies. Asterisk indicates nonspecific bands. (g) DNA content was determined by flow cytometry at 72 h after transfection (n = 3). Error bars indicate mean ± SD. (h) Cell lysates were prepared at 72 h after transfection and were immunoblotted using the indicated antibodies.
Depleting the rRNA processing factors results in the accumulation of p53 and G1 cell cycle arrest, which are RPL11 dependent and MYBPB1A independent
Defects in rRNA processing by the depletion of rRNA processing factors, such as PES1, WDR3 and Las1L, reportedly results in the accumulation of p53 and cell cycle arrest at the G1 phase52,53,54. In addition, we have recently found that persistent depletion of several rRNA processing factors induces p53-dependent cellular senescence46. To assess the mechanistic difference between impaired rRNA transcription- and impaired rRNA processing-induced p53 activation, we depleted several rRNA processing factors in MCF-7 (Figs. 4 and S7) and in U2OS cells (Fig. S8). We first confirmed that siRNA treatment efficiently reduced these factors (Fig. 4a,b) and induced defects in rRNA processing (Fig. 4c). Depleting rRNA processing factors caused p53 accumulation, p53-dependent p21 induction (Figs. 4d and S8a) and p53-dependent cell cycle arrest at the G1 phase, as shown by FACS analysis (Figs. 4e, S7 and S8b). Consistent with FACS analysis, immunoblotting showed that Cyclin D1, a marker of the G1 phase, increased and Cyclin A, which accumulates in the S, the G2 and the early M phases, decreased by depleting these factors (Figs. 4f and S8c).
Depleting rRNA processing factors causes RPL11-dependent and Myb-binding protein 1A (MYBBP1A)-independent p53 activation.
(a–c) MCF-7 cells were transfected with siCtrl, siPES1, siWDR3, siNOL1 or siUTP6. (a) Expression of rRNA processing factors was assessed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) at 48 h after transfection (n = 3). Error bars indicate mean ± SD. (b) Protein levels of rRNA processing factors were assessed by immunoblotting at 48 h after transfection. (c) Northern blotting was performed at 60 h after transfection using a probe specific for the rRNA region. An ethidium bromide-stained gel is shown at the bottom as the loading control. (d–g) MCF-7 cells were transfected with combinations of siRNAs as indicated. (d) Cell lysates were prepared at 60 h after transfection and immunoblotted using the indicated antibodies. Lysates from 2 μg/ml camptothecin (CPT)-treated cells were used as the positive control. (e) DNA content was determined by flow cytometry at 72 h after transfection (n = 3). Error bars indicate mean ± standard deviation (SD). (f) Cell lysates were prepared at 72 h after transfection and immunoblotted using the indicated antibodies. (g) Cell lysates were prepared at 60 h after transfection and immunoblotted using the indicated antibodies. Lysates from 2 μg/ml CPT-treated cells were used as the positive control. (h) Nucleolar RNA content was spectrophotometrically quantified at 60 h after transfection (n = 3). Nucleolar RNA content of control cells was normalised to 100%. Error bars indicate mean ± SD. (i) Immunofluorescence staining was performed using the indicated antibodies at 60 h after transfection.
Depleting the processing factors scarcely induced K382-acetylation of p53, expression of BAX and apoptosis (Figs. 4d,e, S8a and S8b), which differed from depleting the rRNA transcription factors (Figs. 1, S2–S6). Therefore, we depleted either RPL11 or MYBBP1A with the rRNA processing factors to identify the protein that activates p53 when rRNA processing was inhibited. We found that depleting RPL11 (but not MYBBP1A) compromised the accumulation of p53 and the induction of p21 (Figs. 4g and S8d). This result shows that RPL11 is involved in activating p53 caused by impaired rRNA processing, whereas MYBBP1A is not involved. Consistent with this result, when the rRNA processing factors were depleted, nucleolar RNA content did not drop (Figs. 4h and S8e)46 and MYBBP1A was not translocated to the nucleoplasm (Figs. 4i and S8f).
Next, we depleted 10 other rRNA processing factors in MCF-7 cells. Similar cellular responses, such as retention of MYBBP1A in the nucleolus (Fig. 5a), p53 accumulation (Fig. 5b) and G1 cell cycle arrest (Figs. 5c,d and S9), were observed in these cells, indicating a relationship between MYBBP1A-independent p53 activation and G1 cell cycle arrest. Taken together, these results show that defects in rRNA processing result in the accumulation of p53 and p21 and cell cycle arrest at the G1 phase, which depends on RPL11.
Depletion of various rRNA processing factors activates p53 without translocating Myb-binding protein 1A (MYBBP1A).
(a–d) MCF-7 cells were transfected with siRNAs against rRNA processing factors. (a) Immunofluorescence staining was performed using the indicated antibodies at 60 h after transfection. (b) Cell lysates were prepared at 60 h after transfection and immunoblotted using the indicated antibodies. Lysates from 2 μg/ml camptothecin-treated cells were used as the positive control. (c) DNA content was determined by flow cytometry at 72 h after transfection (n = 3). Error bars indicate mean ± standard deviation. (d) Cell lysates were prepared at 72 h after transfection and immunoblotted using the indicated antibodies.
Finally, we performed a chromatin immunoprecipitation (ChIP) assay to determine why inhibiting rRNA transcription and rRNA processing caused apoptosis and cell cycle arrest. We noticed that p53 bound to the p21 promoter when either TIF-IA, an rRNA transcription factor, or PES1, an rRNA processing factor, were depleted (Fig. 6a). In contrast, p53 bound to the BAX promoter when TIF-IA, but not PES1, was depleted (Fig. 6b). These results indicate that MYBBP1A-mediated K382 acetylation of p53 enhances p53 promoter-binding activity and causes apoptosis when rRNA transcription is inhibited.
Inhibiting rRNA transcription enhances p53 DNA-binding activity.
MCF-7 cells were transfected with siCtrl, siPES1 or siTIF-IA. The chromatin immunoprecipitation assay was performed using normal mouse IgG and anti-p53 antibodies. The p53-binding regions of the p21 promoter (a) or the BAX promoter (b) were amplified and analysed by reverse transcription-quantitative polymerase chain reaction (n = 3). Error bars indicate mean ± standard deviation.
Discussion
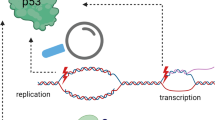
The present data demonstrate that the nucleolus strictly regulates the activation of p53 and subsequent cell fate. Inhibiting rRNA transcription enhanced p53 acetylation and led to apoptosis, whereas inhibiting rRNA processing led to the accumulation of p53 and caused cell cycle arrest at the G1 phase. The nucleolar protein MYBBP1A is indispensable for the former process but not for the latter process (Fig. 7).
Inhibition of rRNA transcription causes p53 activation and apoptosis42,47. When rRNA transcription was inhibited by depleting rRNA transcription factors, nucleolar RNA content was decreased and the nucleolus disrupted (Figs. 2,3, S3, S5 and S6). Nucleolar RNA content was decreased in a stepwise manner either by depleting several rRNA transcription factors (Figs. 2 and S3) or by depleting an identical rRNA transcription factor (TIF-IA) with various siRNA concentrations (Figs. 3, S5 and S6). In both cases, the reduced nucleolar RNA content was correlated with the MYBBP1A translocation to the nucleoplasm (Figs. 2,3, S3, S5 and S6). Furthermore, the ratio of translocation of MYBBP1A parallelled the upregulation of p53 K382-acetylation (Figs. 2,3, S3, S5 and S6). These results strongly support our previous study47,48, in which we proposed that MYBBP1A is anchored in the nucleolus by binding to nucleolar RNA and that translocated MYBBP1A enhances p53 acetylation by stabilising the binding between p300 and p53.
The data in Figs. 2c,3f, S5d and S6d show that depletion of rRNA transcription factors reduces the protein levels of MYBBP1A, which apparently opposes the increased p53 acetylation (because MYBBP1A enhances p53 acetylation). However, our previous publication may account for the discrepancy between the decreased MYBBP1A and the enhanced p53 acetylation as follows: in response to nucleolar disruption, MYBBP1A translocated to the nucleoplasm, where it bound to nonacetylated p53, enhanced its acetylation and then dissociated from acetylated p5350. Thus, persistent interaction between MYBBP1A and p53 would not be required for p53 acetylation: MYBBP1A may be degraded after it transiently associates with p53 and induces p53 acetylation. This idea is supported by the result that MG132 alleviated the reduction in MYBBP1A protein levels by siTIF-IA treatment (Fig. S10). Analysing the significance of MYBBP1A degradation and identification of ubiquitin ligases that degrade MYBBP1A may be a topic of interest for future studies.
We have very recently found that persistent depletion of rRNA processing factors, which mimics nucleolar status under replicative stress, leads to cellular senescence46. When rRNA processing was inhibited, nucleolar RNA content rather increased (Figs. 4h and S8e) and the nucleolus was enlarged (Figs. 4i,5a and S8e)46. Consistent with this result, MYBBP1A did not translocate from the nucleolus (Figs. 4i,5a and S8f). In this case, the nonacetylated form of p53 increased and p21 (but not BAX) accumulated (Figs. 4d,5b and S8a), which was independent of MYBBP1A but dependent on RPL11 (Figs. 4g and S8d). Depletion of almost all of the rRNA processing factors that we tested increased G1 phase cells without affecting the ratio of subG1 phase cells (Figs. 4e,5c and S8b), which would be a prerequisite for the induction of cellular senescence.
Post-translational modifications of p53, such as acetylation and phosphorylation, regulate sequence-specific DNA binding of p531,4,5,6,7. Acetylation of p53 at the C-terminal region reportedly strengthens its DNA-binding activity17,18. Moreover, the strength of p53 DNA-binding activity determines promoter selectivity16,55,56. These observation suggest that nonacetylated p53 possesses weak DNA-binding activity and binds to REs on cell cycle genes, such as the p21 gene, whereas acetylation of p53 at the C-terminal region confers enhanced DNA-binding activity, which leads to p53 binding to REs on proapoptotic targets, including BAX, NOXA and PUMA. Consistent with these observations, we showed that inhibiting rRNA transcription induced apoptosis via MYBBP1A by enhancing p53 K382-acetylation. To support this model, acetylated p53 robustly induced BAX expression (compare lane 5 with lanes 6 or 7; Fig. S11) even when p53 protein levels were similar, showing that p53 acetylation status influences BAX expression. However, as reported in the previous publication26, we still cannot exclude the effect of p53 accumulation on BAX expression from the result of Fig. 2c, in which BAX expression appears to be linked to protein levels of p53.
Finally, we propose that the nucleolus functions as a sensor that recognises the strength of cellular stress. Various types of stress, including anticancer drugs, gamma irradiation and glucose starvation, inhibit rRNA transcription48,51,57,58. These stressors inhibit rRNA transcription in a strength-dependent manner. Here we showed that MYBBP1A translocated from the nucleolus to the nucleoplasm in parallel with the inhibition of rRNA transcription (Figs. 1, S3, S5 and S6). Taken together, these results suggest that the nucleolus functions as a sensor that monitors the strength of cellular stress and induces MYBBP1A-dependent p53 acetylation and apoptosis by regulating the degree of rRNA transcription. Several compounds inhibit rRNA processing at a low dose but inhibit rRNA transcription at a high dose51, suggesting that lower stress inhibits rRNA processing and higher stress inhibits rRNA transcription. Thus, it is possible that the nucleolus induces cell cycle arrest and cellular senescence under lower stress conditions and induces apoptosis under higher stress conditions, at least in certain settings.
Methods
Cell culture & treatments
MCF-7 human breast cancer cells were maintained in Dulbecco’s modified Eagle’s medium containing 1000 mg/l glucose supplemented with 10% fetal bovine serum, 100 units/ml penicillin and 100 μg/ml streptomycin. Camptothecin (2 mg/ml; Sigma, St. Louis, MO, USA) treatment was generally performed 24 h after cell seeding.
RNA interference
Cells at 30%–50% confluence were transfected with 20 nM siRNA using LipofectAmine RNAi max (Invitrogen, Carlsbad, CA, USA) to transfect the siRNAs, according to the manufacturer’s protocol and were incubated for the indicated times. All siRNAs were purchased from Invitrogen. The siRNA sequences are shown in Supplementary Table S1. The Stealth RNAiTM Luciferase reporter control duplex was used as a negative control.
Antibodies
The following antibodies were used in these experiments: β-actin (Sigma), p53 (DO-1, Santa Cruz Biotechnology, Santa Cruz, CA, USA), acethyl-p53 (lysine382, Cell Signaling Technology, Danvers, MA, USA), p21 (F-5, Santa Cruz Biotechnology), BAX (Abcam, Cambridge, MA, USA), PARP (Cell Signaling Technology), Cyclin A (H-432, Santa Cruz Biotechnology), Cyclin D1 (Cell Signaling Technology), POLR1B (N-17, Santa Cruz Biotechnology), UBF (F-9, Santa Cruz Biotechnology), POLR1A (C-1, Santa Cruz Biotechnology), TIF-IA (C-20, Santa Cruz Biotechnology), PES1 (Bethyl Laboratories, Montgomery, TX, USA), WDR3 (Acris Antibodies, San Diego, CA, USA), NOL1 (Bethyl Laboratories), UTP6 (Gene Tex, Irvine, CA, USA) and RPL11 (3A4A7, Invitrogen). The rabbit anti-MYBBP1A antibody was raised against a synthetic peptide corresponding to the human-MYBBP1A 1265–1328 amino acid sequence.
Immunofluorescent staining
Immunofluorescent staining was performed as described previously48. MCF-7 cells were fixed in 4% paraformaldehyde at room temperature for 15 min and permeabilized with 0.5% Triton X-100 buffer (20 mM HEPES, 150 mM KCl and 0.5% Triton X-100) for 10 min at room temperature. Samples were blocked with 0.5% goat serum in TBS-T buffer (20 mM Tris pH 7.5, 150 mM NaCl and 0.05% Tween-20) for 1 h, washed with phosphate-buffered saline (PBS; 140 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4 and 8.1 mM Na2HPO4) and incubated with the indicated antibodies for 2 h. The samples were washed with PBS and incubated with Alexa Fluor® 594 goat anti-mouse IgG (H + L) (Invitrogen) or Alexa Fluor® 488 goat anti-rabbit IgG (H + L) (Invitrogen) for 1 h. The samples were washed again with PBS, stained with 4, 6-diamidino-2-phenylindole (Dojindo, Rockville, MD, USA) and mounted on slides with Vectashield (Vector Laboratories, Burlingame, CA, USA). The samples were visualized by Biozero immunofluorescence microscopy (Keyence, Osaka, Japan).
Immunoblotting
Cells were harvested by trypsinization, washed with PBS and lysed in RIPA buffer [25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 1% Triton X-100, 1% sodium deoxycholate and 0.1% sodium dodecyl sulfate-polyacrylamide (SDS) plus a protease inhibitor cocktail (Nacalai Tesque, Inc., Kyoto, Japan) for 30 min on ice. The lysates were cleared by centrifugation at 16,100 × g for 30 min at 4 °C. Protein concentrations were evaluated using a BCA kit (Thermo Scientific, Rockford, IL, USA). Extracted proteins were separated by SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (Millipore, Milford, MA, USA). After blocking with 5% skim milk in TBS-T buffer- for 1 h, the membranes were incubated with primary antibodies for at least 1 h. After washing with TBS-T buffer 3 times, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody for 1 h. Then the membrane was washed with TBS-T buffer twice and with TBS once. Bands were detected with Chemi-Lumi One (Nacalai Tesque) or the Immobilon Western blotting detection kit (Millipore).
Nucleoli purification and quantitative determination of nucleolar RNA content
Nucleoli were isolated from 1.2 × 107 MCF-7 cells by density gradient fractionation, as described previously59. Total nucleolar RNA was prepared from the isolated nucleoli and quantified by spectrometry.
Northern blotting
RNA was isolated with a FastPure® RNA kit (Takara Bio) according to the manufacturer’s instructions. Total RNA (2 μg) was separated on a 1% agarose-formaldehyde gel and blotted on nylon membranes (GE Healthcare, Bucks, UK). The following DNA oligonucleotides were 5`end-labeled using T4 polynucleotide kinase in the presence of [g-32P] ATP: 5′-CCTCCGCGCCGGAACGCGCTAGGTACCTGGACGGCGGGGGGGCGGACG-3′ for ITS1. Membranes were preincubated with ULTRAhyb buffer (Ambion, Austin, TX, USA) at 65 °C for 30 min and the radioactively labeled probes were then added overnight. Membranes were shortly washed in 1 × SSC and 0.2 × SSC.
RNA purification and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated with a FastPure® RNA kit (Takara Bio, Shiga, Japan) according to the manufacturer’s instructions. Total RNA (1 μg) was reverse transcribed with the PrimeScript® 1st Strand cDNA Synthesis kit (Takara Bio). The real-time quantitative PCR analysis was performed to amplify fragments representing the indicated mRNA or for pre-rRNA expression using the Thermal Cycle DiceTM TP800 (Takara) and SYBR. Indicated mRNA or pre-rRNA levels were normalised to β-actin mRNA levels. The primer sequences can be found in Supplementary Table S2.
ChIP assay
ChIP assay was performed as described previously60 with minor modifications. DNA was amplified by RT-qPCR as described above. The primers for real-time PCR were p53-binding site in the human p21 gene promoter region forward primer: 5′-GTGGCTCTGATTGGCTTTCTG-3′, human p21 gene reverse primer: 5′-CTTGGGCTGCCTGTTTTCAG-3′; human BAX gene forward primer: 5′-TAATCCCAGCGCTTTGGAAG-3′; and the human BAX gene reverse primer: 5′-TTGCTAGATCCAGGTCTCTGCA-3′. Samples were normalised based on the amount of inputted DNA.
Cell cycle analysis
The cell cycle was analysed as described previously48. The cells were detached with trypsin, collected with the supernatant and washed twice with ice-cold PBS. The cells were fixed in 75% (v/v) cold ethanol for no less than 12 h and washed in ice-cold PBS. After removing the supernatant, the cells were stained with the Guava® Cell Cycle Reagent (Millipore). Cell cycle populations were determined using the Guava® EasyCyte system (Guava Technologies, Hayward, CA, USA), according to the manufacturer’s recommendations. Guava® Cell Cycle software (Guava Technologies) was used to determine the different cell cycle phases in the cell populations.
Additional Information
How to cite this article: Kumazawa, T. et al. Gradual reduction in rRNA transcription triggers p53 acetylation and apoptosis via MYBBP1A. Sci. Rep. 5, 10854; doi: 10.1038/srep10854 (2015).
References
Kruse, J. P. & Gu, W. Modes of p53 regulation. Cell 137, 609–622 (2009).
Lee, J. T. & Gu, W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 17, 86–92 (2010).
Olson, M. O. Sensing cellular stress: another new function for the nucleolus? Sci. STKE 2004, pe10 (2004).
Appella, E. & Anderson, C. W. Post-translational modifications and activation of p53 by genotoxic stresses. FEBS 268, 2764–2772 (2001).
Bode, A. M. & Dong, Z. Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer 4, 793–805 (2004).
Brooks, C. L. & Gu, W. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr. Opin. Cell Biol. 15, 164–171 (2003).
Carter, S. & Vousden, K. H. Modifications of p53: competing for the lysines. Curr. Opin. Genet. Dev. 19, 18–24 (2009).
Vousden, K. H. & Lane, D. P. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 8, 275–283 (2007).
Harper, J. W., Adami, G. R., Wei, N., Keyomarsi, K. & Elledge, S. J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816 (1993).
el-Deiry, W. S. et al. WAF1, a potential mediator of p53 tumor suppression. Cell 75, 817–825 (1993).
Nakano, K. & Vousden, K. H. PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell 7, 683–694 (2001).
Pawlowski, J. & Kraft, A. S. Bax-induced apoptotic cell death. Proc. Natl. Acad. Sci. U.S.A. 97, 529–531 (2000).
Oda, E. et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288, 1053–1058 (2000).
Oda, K. et al. p53AIP1, a potential mediator of p53-dependent apoptosis and its regulation by Ser-46-phosphorylated p53. Cell 102, 849–862 (2000).
Contente, A., Dittmer, A., Koch, M. C., Roth, J. & Dobbelstein, M. A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat. Genet. 30, 315–320 (2002).
Weinberg, R. L., Veprintsev, D. B., Bycroft, M. & Fersht, A. R. Comparative binding of p53 to its promoter and DNA recognition elements. J. Mol. Biol. 348, 589–596 (2005).
Gu, W. & Roeder, R. G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595–606 (1997).
Luo, J. et al. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 101, 2259–2264 (2004).
Tang, Y., Luo, J., Zhang, W. & Gu, W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 24, 827–839 (2006).
Sykes, S. M. et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 24, 841–851 (2006).
Taira, N., Nihira, K., Yamaguchi, T., Miki, Y. & Yoshida, K. DYRK2 is targeted to the nucleus and controls p53 via Ser46 phosphorylation in the apoptotic response to DNA damage. Mol. Cell 25, 725–738 (2007).
D’Orazi, G. et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat. Cell Biol. 4, 11–19 (2002).
Beckerman, R. & Prives, C. Transcriptional regulation by p53. Cold Spring Harb. Perspect. Biol. 2, a000935 (2010).
Espinosa, J. M. Mechanisms of regulatory diversity within the p53 transcriptional network. Oncogene 27, 4013–4023 (2008).
Vousden, K. H. & Prives, C. Blinded by the Light: The Growing Complexity of p53. Cell 137, 413–431 (2009).
Lai, P. B., Chi, T. Y. & Chen, G. G. Different levels of p53 induced either apoptosis or cell cycle arrest in a doxycycline-regulated hepatocellular carcinoma cell line in vitro. Apoptosis 12, 387–393 (2007).
Tanaka, T., Ohkubo, S., Tatsuno, I. & Prives, C. hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell 130, 638–650 (2007).
Boulon, S., Westman, B. J., Hutten, S., Boisvert, F. M. & Lamond, A. I. The nucleolus under stress. Mol. Cell 40, 216–227 (2010).
Rubbi, C. P. & Milner, J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 22, 6068–6077 (2003).
Zhang, Y. & Lu, H. Signaling to p53: ribosomal proteins find their way. Cancer Cell 16, 369–377 (2009).
Lohrum, M. A., Ludwig, R. L., Kubbutat, M. H., Hanlon, M. & Vousden, K. H. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3, 577–587 (2003).
Marechal, V., Elenbaas, B., Piette, J., Nicolas, J. C. & Levine, A. J. The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol. Cell. Biol. 14, 7414–7420 (1994).
Kamijo, T. et al. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl. Acad. Sci. U.S.A. 95, 8292–8297 (1998).
Pomerantz, J. et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell 92, 713–723 (1998).
Dai, M. S. & Lu, H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 279, 44475–44482 (2004).
Dai, M. S. et al. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell. Biol. 24, 7654–7668 (2004).
Jin, A., Itahana, K., O’Keefe, K. & Zhang, Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell. Biol. 24, 7669–7680 (2004).
Kurki, S. et al. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell 5, 465–475 (2004).
Saxena, A., Rorie, C. J., Dimitrova, D., Daniely, Y. & Borowiec, J. A. Nucleolin inhibits Hdm2 by multiple pathways leading to p53 stabilization. Oncogene 25, 7274–7288 (2006).
Chen, Z., Knutson, E., Wang, S., Martinez, L. A. & Albrecht, T. Stabilization of p53 in human cytomegalovirus-initiated cells is associated with sequestration of HDM2 and decreased p53 ubiquitination. J. Biol. Chem. 282, 29284–29295 (2007).
Zhu, Y. et al. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol. Cell 35, 316–326 (2009).
Yuan, X. et al. Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest and p53-mediated apoptosis. Mol. Cell 19, 77–87 (2005).
Donati, G., Peddigari, S., Mercer, C. A. & Thomas, G. 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep. 4, 87–98 (2013).
Li, M. & Gu, W. A critical role for noncoding 5S rRNA in regulating Mdmx stability. Mol. Cell 43, 1023–1032 (2011).
Sloan, K. E., Bohnsack, M. T. & Watkins, N. J. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep. 5, 237–247 (2013).
Nishimura, K. et al. Perturbation of Ribosome Biogenesis Drives Cells into Senescence through 5S RNP-Mediated p53 Activation. Cell Rep. 10, 1310–1323 (2015).
Kuroda, T. et al. RNA content in the nucleolus alters p53 acetylation via MYBBP1A. EMBO J. 30, 1054–1066 (2011).
Kumazawa, T. et al. Novel nucleolar pathway connecting intracellular energy status with p53 activation. J. Biol. Chem. 286, 20861–20869 (2011).
Ono, W. et al. Nucleolar protein, Myb-binding protein 1A, specifically binds to nonacetylated p53 and efficiently promotes transcriptional activation. Biochem. Biophys. Res. Commun. 434, 659–663 (2013).
Ono, W. et al. The nucleolar protein Myb-binding protein 1A (MYBBP1A) enhances p53 tetramerization and acetylation in response to nucleolar disruption. J. Biol. Chem. 289, 4928–4940 (2014).
Burger, K. et al. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J. Biol. Chem. 285, 12416–12425 (2010).
Holzel, M. et al. Defects in 18 S or 28 S rRNA processing activate the p53 pathway. J. Biol. Chem. 285, 6364–6370 (2010).
McMahon, M., Ayllon, V., Panov, K. I. & O’Connor, R. Ribosomal 18 S RNA processing by the IGF-I-responsive WDR3 protein is integrated with p53 function in cancer cell proliferation. J. Biol. Chem. 285, 18309–18318 (2010).
Castle, C. D., Cassimere, E. K., Lee, J. & Denicourt, C. Las1L is a nucleolar protein required for cell proliferation and ribosome biogenesis. Mol. Cell. Biol. 30, 4404–4414 (2010).
Schlereth, K. et al. DNA binding cooperativity of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 38, 356–368 (2010).
Schlereth, K., Charles, J. P., Bretz, A. C. & Stiewe, T. Life or death: p53-induced apoptosis requires DNA binding cooperativity. Cell Cycle 9, 4068–4076 (2010).
Hoppe, S. et al. AMP-activated protein kinase adapts rRNA synthesis to cellular energy supply. Proc. Natl. Acad. Sci. U.S.A. 106, 17781–17786 (2009).
Kruhlak, M. et al. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature 447, 730–734 (2007).
Andersen, J. S. et al. Nucleolar proteome dynamics. Nature 433, 77–83 (2005).
Kaeser, M. D. & Iggo, R. D. Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc. Natl. Acad. Sci. U.S.A. 99, 95–100 (2002).
Acknowledgements
We thank enago for the English language review. This work was supported by the Reserch Fellowship (to T.K. and K.N.) from the Japan Society for the Promotion of Science (JSPS).
Author information
Authors and Affiliations
Contributions
T.K., K.N. and K.K. conceived the project and designed experiments. T.K., K.N. and K.K. wrote the manuscript. T.K., K.N., N.K., S.H. and Y.H. performed experiments. T.K. and K.N. analyzed the data with significant assistance from N.K., S.H. and Y.H.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Kumazawa, T., Nishimura, K., Katagiri, N. et al. Gradual reduction in rRNA transcription triggers p53 acetylation and apoptosis via MYBBP1A. Sci Rep 5, 10854 (2015). https://doi.org/10.1038/srep10854
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep10854
- Springer Nature Limited
This article is cited by
-
NAT10-mediated upregulation of GAS5 facilitates immune cell infiltration in non-small cell lung cancer via the MYBBP1A-p53/IRF1/type I interferon signaling axis
Cell Death Discovery (2024)
-
MiR-330-5p and miR-1270 target essential components of RNA polymerase I transcription and exhibit a novel tumor suppressor role in lung adenocarcinoma
Cancer Gene Therapy (2023)
-
rDNA Transcription in Developmental Diseases and Stem Cells
Stem Cell Reviews and Reports (2023)
-
p38-MAPK-mediated translation regulation during early blastocyst development is required for primitive endoderm differentiation in mice
Communications Biology (2021)
-
HMGB proteins involved in TOR signaling as general regulators of cell growth by controlling ribosome biogenesis
Current Genetics (2018)