Abstract
Renal fibrosis is the final common pathway leading to decreased renal function. No therapy has been established to prevent it. In order to establish a therapeutic approach and target molecule for renal fibrosis, we investigated the effects of Smad4 knockdown by siRNAs on renal fibrosis in vivo. Renal fibrosis mice were produced by single intraperitoneal injection of folic acid. siRNAs targeted to Smad4 (Smad4-siRNAs) (5 nmol) were injected into each mouse by systemic tail vein injection three times per week. Non-targeted siRNAs (control-siRNAs) were injected in the same way for a control group. The siRNAs were delivered to the interstitial fibrous area and tubules. Smad4-siRNAs significantly knocked down Smad4 expression and inhibited renal fibrosis. They also inhibited α-SMA-positive myofibroblasts. Control-siRNAs did not show these effects. The results of this study suggest that Smad4 knockdown is one of the crucial therapeutic options for the prevention of renal fibrosis in vivo.
Similar content being viewed by others
Introduction
Renal tubulointerstitial fibrosis, also called renal fibrosis, is the final common pathway leading to decreased renal function1,2,3. It leads to end-stage renal disease regardless of the underlying etiology in all progressive chronic kidney diseases1,2,3. It is characterized by the proliferation of myofibroblasts and the excess accumulation of extracellular matrix as collagens in interstitial space4. No therapy has been established to prevent renal fibrosis. Since many different kinds of cell, such as immune cells, vascular endothelial cells, tubular epithelial cells and fibroblasts, have been considered to contribute to the development of renal fibrosis, the investigation of therapeutic approaches, including the exploration of target genes, for the inhibition of renal fibrosis in vivo is crucially important to improve the prognosis of all chronic kidney diseases5,6,7,8,9,10,11,12.
RNA interference (RNAi) is a sequence-specific gene-silencing mechanism13,14. It has evolved from a valuable research tool for exploring target genes towards a potentially powerful therapeutic approach for various diseases15,16,17. The use of synthetic small interfering RNAs (siRNAs) for gene silencing by the RNAi mechanism eliminates safety concerns associated with viral vectors. Furthermore, siRNAs, which enter the late stage of the endogenous RNAi pathway, are less likely to interfere with the gene regulation carried out by the endogenous microRNA machinery18. Several studies have reported the effects of gene silencing by siRNAs for the exploration of target genes and for the treatment of renal diseases, including acute kidney injury, in an experimental kidney transplantation model and for glomerular disease19,20,21,22; however, only a few studies have reported the validity of target gene silencing by siRNAs for renal fibrosis in vivo21.
TGF-β1-Smad has been reported as the main signaling pathway that regulates renal fibrosis23,24. Receptor-activated Smad (Smad2 and Smad3) combines with a common Smad called Smad4 and then this complex translocates to the nucleus, where it binds to DNA elements to promote the transcription of various genes that regulate renal fibrosis23,24. These lines of evidence suggest that Smad4 may be a key molecule that can regulate renal fibrosis and the proliferation of myofibroblasts; however, the effects of Smad4 inhibition in the kidney in vivo on renal fibrosis have not been fully clarified.
In the present study, we investigated the effects of Smad4 knockdown on renal fibrosis and the expression of myofibroblasts in vivo by tail vein injection of siRNAs targeting Smad4 to kidney.
Results
The distribution of siRNAs systemically delivered by tail vein injection in kidney
The siRNAs (red) systemically delivered by tail vein injection were largely distributed in renal tubules (Figure 1). Co-staining with a renal tubular cell marker, fluorescein-labeled lotus tetragonolobus lectin (green), showed that the siRNAs (red) were also distributed in the interstitial fibrous area where the renal tubular cells did not have a regular form in renal fibrosis mice (Figure 1: arrow).
The distribution of systemically delivered siRNAs in kidney.
Cy3-labeled siRNAs that were systemically delivered by tail vein injection were detected in most tubular cells and the interstitial fibrous area (arrow), in the kidney. No positivity was observed upon control (vehicle) injection. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; LTL, lotus tetragonolobus lectin.
The knockdown of Smad4 of the kidney by systemically delivered Smad4-siRNAs
The results of qRT-PCR, western blot analysis and immunofluorescence showed that Smad4 expression in the kidney was increased in the renal fibrosis mice compared with that in mock mice. Systemically delivered Smad4-siRNAs significantly knocked down the increased Smad4 expression in the kidney; however, Control-siRNAs did not knock it down in renal fibrosis mice in vivo (Figures 2 and 3).
The results of qRT-PCR and western blot analysis for the knockdown of Smad4 in the kidney by systemically delivered Smad4-siRNAs in renal fibrosis.
qRT-PCR (A) and western blot analysis (B) of Smad4 expression in each group. Each group: n = 6, values are mean ± standard error (error bars) of at least three independent experiments. Abbreviation: NS, not significant; *, p <0.05.
The results of immunofluorescence staining for the knockdown of Smad4 in the kidney by systemically delivered Smad4-siRNAs in renal fibrosis.
A. Representative immunofluorescence staining of kidney sections in each group. B. The quantitative analysis of Smad4-positive cells in each group. Each group: n = 6, values are mean ± standard error (error bars) of at least three independent experiments. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; NS, not significant; *, p <0.05.
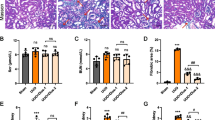
The effects of systemically delivered Smad4-siRNAs on kidney weights and renal functions in renal fibrosis
The bilateral kidney weights of each group were as follows: mock, 380.8 ± 8.05 mg; renal fibrosis, 314.4 ± 14.4 mg; renal fibrosis + Smad4-siRNAs, 360.0 ± 34.6 mg; and renal fibrosis + Control-siRNAs, 290.0 ± 13.3 mg. The kidney weights of renal fibrosis mice were significantly lower than those of mock mice (p <0.05). Systemically delivered Smad4-siRNAs significantly inhibited the decreased kidney weights (p <0.05); however, Control-siRNAs did not (NS). The serum blood urea nitrogen (BUN) and creatinine (Cr) levels of each group were as follows [BUN/Cr (mg/dl)]: mock, 28.3 ± 0.33/0.10 ± 0.00 mg/dl; renal fibrosis, 35.7 ± 1.87/0.157 ± 0.02 mg/dl; renal fibrosis + Smad4-siRNAs, 27.0 ± 4.58/0.10 ± 0.00 mg/dl; and renal fibrosis + Control-siRNAs, 56.0 ± 6.34/0.23 ± 0.05 mg/dl. The serum BUN and Cr levels showed trends of increases in the renal fibrosis mice and they seemed to be decreased by systemically delivered Smad4-siRNAs. On the other hand, they seemed to be further increased by Control-siRNAs compared with those of renal fibrosis mice without siRNA injection: however, their change did not reach statistical significance.
The inhibition of renal fibrosis by systemically delivered Smad4-siRNAs
The interstitial fibrosis changes were histologically detected in renal fibrosis mice by Azan staining and Sirius red staining (Figures 4A and 4B). The areas with positive blue staining showed collagen fibers on Azan staining (Figure 4A). The positive blue-stained areas increased in renal fibrosis mice compared with those in mock mice (Figure 4A). These fibrosis areas were inhibited by systemically delivered Smad4-siRNAs; however, they were not inhibited by Control-siRNAs (Figure 4A). The quantified levels of fibrosis areas evaluated by Sirius red staining in each group were as follows: mock, 3.35 ± 0.32%/field; renal fibrosis, 15.5 ± 2.83%/field; renal fibrosis + Smad4-siRNAs, 8.60 ± 0.98%/field; and renal fibrosis + Control-siRNAs, 14.42 ± 1.45%/field (Figure 4B). The increased fibrous changes in renal fibrosis mice were significantly inhibited by systemically delivered Smad4-siRNAs; however, they were not inhibited by control-siRNAs (Figure 4B).
The effects of systemically delivered Smad4-siRNAs on fibrotic change in kidney.
A. Representative Azan staining of kidney sections in each group. The areas of positive blue staining showed collagen fibers. B. The quantitative analysis of fibrotic areas in each group. Each group: n = 6, values are mean ± standard error (error bars). Abbreviations: NS, not significant; *, p <0.05; **, p <0.01.
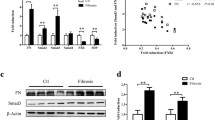
The effects of systemically delivered Smad4-siRNAs on fibrosis markers
Western blot analysis showed that the expression of α-SMA in the kidney was increased in renal fibrosis mice compared with that in mock mice (Figure 5A). This increase was significantly inhibited by systemically delivered Smad4-siRNAs; however, it was not inhibited by Control-siRNAs (Figure 5A). Immunofluorescence also showed that the expression of α-SMA-positive myofibroblasts was significantly increased in renal fibrosis mice (Figure 5B). This increase was significantly inhibited by systemically delivered Smad4-siRNAs; however, it was not inhibited by Control-siRNAs (Figure 5B). qRT-PCR showed that the expression of fibroblast-specific protein 1 (FSP-1) was increased in renal fibrosis mice compared with that in mock mice (Figure 6). This increase was significantly inhibited by systemically delivered Smad4-siRNAs; however, it was not inhibited by Control-siRNAs (Figure 6). The expression of collagen 1A2 also showed a similar trend to FSP-1; however, it did not reach statistical significance (Figure 6).
The effects of systemically delivered Smad4-siRNAs on the expression of α-SMA and α-SMA-positive myofibroblasts.
Western blot analysis of α-SMA expression (A) and representative immunofluorescence and quantitative analysis (B) of α-SMA-positive myofibroblasts in each group. Each group: n = 6, values are mean ± standard error (error bars). Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; α-SMA, α-smooth muscle actin; NS, not significant; *, p <0.05.
The effects of systemically delivered Smad4-siRNAs on fibrotic markers.
The changes of FSP1 and collagen1A2 in each group investigated by qRT-PCR. Each group: n = 6, values are mean ± standard error (error bars). Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; FSP-1, fibroblast-specific protein 1; NS, not significant; *, p <0.05.
The changes of signal conductance in renal fibrosis by systemically delivered Smad4-siRNAs
We investigated the changes in the expression of transforming growth factor-β1 (TGF-β1), Snail1, Slug, matrix metalloproteinase-2 (MMP-2), vascular endothelial growth factor (VEGF) and Serpine1, which have been considered to be important signaling molecules for the development of renal fibrosis. Among them, the expression of Serpine1 was increased in renal fibrosis mice compared with that in mock mice (Figure 7). This increase was significantly inhibited by systemically delivered Smad4-siRNAs; however, it was not inhibited by Control-siRNAs (Figure 7). The expression levels of TGF-β1, Snail1, Slug, MMP-2 and VEGF also showed similar trends to Serpine1; however, they did not reach statistical significance (Figure 7).
The effects of systemically delivered smad4-siRNAs on signal conductance in renal fibrosis.
The changes of TGF-β1, Snail1, Slug, MMP-2, VEGF and Serpine1 in each group investigated by qRT-PCR. Each group: n = 6, values are means ± standard error (error bars). Abbreviations: TGF-β1, transforming growth factor-β; MMP-2, matrix metalloproteinase2; VEGF, vascular endothelial growth factor. *, p <0.05.
Discussion
The results of this study show that systemic Smad4-siRNA delivery by tail vein injection prevents renal fibrosis by inhibiting the increased expression of myofibroblasts. These results suggest that Smad4 has pivotal roles in the expression of myofibroblasts resulting in the development of renal fibrosis and it would be one of the crucial therapeutic targets for the inhibition of renal fibrosis. Renal fibrosis is a common pathway in all progressive chronic kidney diseases, regardless of the underlying etiology1,2,3. It was reported to be a better predictor of decreased renal function than glomerular lesions1,2,3. Therefore, the inhibition of renal fibrosis is very important to improve the prognosis of all chronic kidney disease patients. Since no specific therapy that can inhibit renal fibrosis has been established, the investigation and development of therapeutic approaches as well as the exploration of target molecules to inhibit renal fibrosis in vivo are very significant. RNAi by siRNAs has evolved from a valuable research tool for exploring target genes towards a potentially powerful therapeutic approach for various diseases15,16,17. Previous studies reported that siRNA-mediated target gene knockdown showed beneficial effects in several renal diseases21,22,25,26. Hamar et al. reported that the hydrodynamic systemic delivery of siRNA targeting Fas protected mice against acute renal failure25. Ma et al. reported that the hydrodynamic systemic delivery of siRNA targeting hypoxia-inducible factor 1 alpha protected against renal ischemic-reperfusion injury26. In addition, Luo et al. reported that the systemic delivery of siRNA targeting connective tissue growth factor (CTGF) by hydrodynamic injection significantly knocked down the expression of CTGF in a chronic allograft nephropathy model21. These lines of evidence show that the hydrodynamic injection method can deliver siRNA to kidney to knock down target genes. Since the hydrodynamic injection method requires quite a large volume of solution and cellular toxicity may occur due to the high-pressure injection27, repetition of this method in each mouse many times seems to be technically difficult and may damage tissues. Therefore, this method may not be applicable for kidney diseases that show chronic and slow progression, such as renal fibrosis. Besides the hydrodynamic injection method, standard intravenous injection that requires 100–200 µl of siRNA solution usually via the tail vein can also deliver siRNA to kidney22,28,29. Molitolis et al. reported that intravenously injected synthetic siRNA targeting p53 protected both the function of proximal tubular cells and renal function after ischemic injury22. Water et al. reported that Mrp-2 siRNA intravenously injected via the tail functionally knocked down Mrp2 in kidney29. In the present study, we also detected that siRNAs systemically administered by tail vein injection were delivered in the interstitial fibrous area as well as renal tubular cells. We also found that the systemic delivery of Smad4-siRNAs by tail vein injection significantly knocked down Smad4 expression and prevented renal fibrosis in vivo. These results suggest that standard intravenous injection of siRNAs may be a good therapeutic approach and enable exploration of target genes in chronic kidney disease; in addition, Smad4 appears to be one of the crucial therapeutic targets for the prevention of renal fibrosis in vivo.
The pathology of renal fibrosis has been characterized by the proliferation of myofibroblasts that produce extracellular matrix in the interstitial space of the kidney4. Previous studies reported epithelial cells, endothelial cells, bone marrow-derived cells and pericytes as the origins of these myofibroblasts, in addition to the resident myofibroblasts in renal fibrosis5,6,7,8,9,10,11,12. However, the obtained results on the origin of myofibroblasts have not necessarily been consistent with each other. Recently, LeBleu et al. reported that 50% of myofibroblasts are local resident fibroblasts, 35% of them are derived from bone marrow cells, 10% of them are derived from endothelial cells and 5% of them are derived from epithelial cells, in a renal fibrosis mouse model induced by unilateral ureteral obstruction (UUO), using genetically engineered mice to track, fate-map and ablate cells12. On the other hand, Humphreys et al. reported that the majority of myofibroblasts were derived from pericytes and those from epithelial cells were not observed in renal fibrosis in UUO model mice by fate-tracing5. As described above, the origin of myofibroblasts is still being explored and the evidence is being accumulated using convincing experimental technology such as fate-tracing with genetically engineered mice in fibrosis model animals. Further studies will need to clarify the origins of the myofibroblasts in which differentiation or proliferation was inhibited by Smad4 knockdown by Smad4-siRNAs in the present study using those genetically engineered mouse models5,12. Recently, Meng et al. reported that disruption of Smad4 from the kidney inhibited renal fibrosis in conditional Smad4 knockout mice30. They also reported that Smad4 knockout fibroblasts from kidney inhibited collagen matrix expression30. In the present study, systemic Smad4-siRNA delivery inhibited Smad4 expression and resulted in the suppression of α-SMA-positive myofibroblasts.
Taking these results together with those of previous studies, Smad4 inhibition could be one of the crucial therapeutic approaches to inhibit the expression of myofibroblasts resulting in renal fibrosis in vivo.
In conclusion, the results of the present study show that Smad4 knockdown by the systemic delivery of Smad4-siRNAs by intravenous injection prevented renal fibrosis by inhibiting the increased expression of α-SMA-positive myofibroblasts. These results suggest that Smad4 is one of the crucial therapeutic targets for the prevention of renal fibrosis in vivo.
Methods
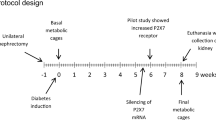
Renal fibrosis mouse model
All animal procedures were approval by the Use and Care of Experimental Animals Committee of the Jichi Medical University Guide for Laboratory Animals and were carried out in accordance with the Jichi Medical University guidelines. C57BL/6 male mice, aged 8 weeks, 20–25 g in body weight, were purchased from CLEA Japan, Inc. (Tokyo, Japan). All mice were housed under antiviral and antibody-free microisolator conditions. To induce renal fibrosis, the mice were intraperitoneally injected with 250 mg/kg folic acid (Sigma-Aldrich, St. Louis, MO, USA) dissolved in phosphate-buffered saline (PBS) (Life Technology Inc., CA, USA) at a concentration of 10 mg/dl one time (day 0). In this mouse model, patchy tubulointerstitial fibrosis occurs within a few weeks after folic acid injection31.
siRNAs
siRNAs targeting Smad4 (Smad4-siRNAs) and universal non-targeted siRNAs (Control-siRNAs) were purchased from Sigma-Aldrich Japan (Ishikari, Hokkaido, Japan). The sequences of the Smad4-siRNAs were as follows: 5′-GAUGAAUUGGAUUCUUUAATT-3′ (sense strand) and 5′-UUAAAGAAUCCAAUUCAUCTT-3′ (antisense strand). The following Control-siRNAs were used: Mission_SIC_001s (sense strand) and Mission_SIC_001as (antisense).
The delivery of siRNAs to the kidney
Smad4-siRNAs (5 nmol) dissolved in 100 μl of DNase/RNase-free distilled water (Life Technology Inc.) were injected via the tail vein into each mouse from the 2nd day after folic acid injection at a frequency of three times per week (renal fibrosis + Smad4-siRNAs). The following groups served as controls: mice that were not injected with folic acid and siRNAs (mock), mice that were injected with folic acid but not siRNAs (renal fibrosis) and mice that were injected with folic acid and Control-siRNAs by the same methods as the Smad4-siRNA group (renal fibrosis + Control-siRNAs). The mice in all groups were sacrificed on the 21st day after folic acid injection. The renal fibrosis + Smad4-siRNA group and the renal fibrosis + Control-siRNA group were injected siRNAs a total of 9 times during the experimental period. Blood samples were collected from the mice from the inferior vena cava and the kidneys were removed after washing by reflux flow of PBS.
qRT-PCR
The kidney specimens were homogenized using a glass homogenizer and a filter column shredder (QIA shredder, Qiagen, Valencia, CA). Then, total RNA from the kidney was isolated using an RNeasy total RNA isolation kit (Qiagen). A total of 1 µg of isolated RNA was reverse-transcribed using the Superscript III first-strand synthesis system (Life Technology) according to the manufacturer's protocol. Real-time RT-PCR was performed using SYBR Green ER qPCR super mix (Life Technology). All reactions were carried out in a 20 µl reaction volume in duplicate. After initial denaturation at 95°C for 10 s, real-time qRT-PCR was performed for 40 cycles under the following conditions: denaturation at 95°C for 15 s and annealing and extension at 60°C for 60 s. Primers for mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Smad4, TGF-β1, Snail1, Slug, MMP-2, VEGF, FSP-1, collagen 1A2 and Serpine1 were purchased from Takara Bio Inc. (Otsu, Shiga, Japan). Quantification of target gene expression was achieved using Step One software (Life Technology). The expression levels of mRNA were normalized with GAPDH used as an endogenous control. Data are expressed as relative quantities compared with the mock group.
Western blot analysis
For western blotting, the kidneys were quickly removed from the mouse, frozen and stored at −80°C prior to use. The kidney samples were homogenized in PRO-PREPTTM protein extraction solution (iNtRON Biotechnology, Inc., Gyeonggi-do, Korea). Then, equal amounts of protein samples (30 μg) were electrophoresed on NuPAGE sodium dodecyl sulfate–polyacrylamide gels (Life Technology) and then transferred onto Immobilon-P membranes (Merck Millipore). After nonspecific protein binding was blocked with PVDF blocking reagent for Can Get Signal (Toyobo, Osaka, Japan), the membrane was incubated overnight at 4°C with an appropriate dilution of the primary antibody [1:100 anti-α-SMA polyclonal antibody (Abcam, Cambridge, England), 1:1000 anti-Smad4 polyclonal antibody (Cell Signaling Technology, MA, USA), 1:5000 anti-GAPDH polyclonal antibody (Santa Cruz Biotechnology)]. After washing, the membrane was incubated for 1 hour at room temperature with a secondary antibody (horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin; GE Healthcare) at a dilution of 1:5000. Then, after washing, the antibody complexes were visualized using the Amersham ECL detection system (GE Healthcare) as directed by the manufacturer. Densitometric analyses for protein quantification were performed using Image J software. The expression levels of α-SMA were normalized with GAPDH used as an endogenous control. Data are expressed as relative quantities compared with the mock group.
Histological analysis
After perfusion with PBS, the kidneys were removed and fixed in 4% paraformaldehyde overnight at 4°C. They were then embedded in paraffin, sectioned and subjected to Azan staining and Sirius red staining to evaluate the intensity of fibrotic changes. Collagen fibers were stained blue by Azan staining and red by Sirius Red staining. For quantitative evaluation of the degree of fibrosis, five fields per section of positive red-stained area by Sirius red staining were analyzed at a magnification of 200-fold. Each analyzed field was chosen randomly and the positive red-stained areas were quantified using computerized image analysis software (Image Pro 5.1, Media Cybernetics Inc., MD, USA).
Fluorescent microscopy
To investigate the distribution of siRNAs that were systemically delivered by the tail vein injection approach in the kidney, Cy-3-labeled siRNAs (3 nmol) (Takara Bio Inc.) were systemically injected via the tail vein into renal fibrosis mice. One hour after injection, the kidneys were removed, cut and fixed at room temperature with 3% paraformaldehyde in 0.1 M cacodylate buffer, pH 7.4, containing 25 mM CaCl2. Fixed tissues were immersed and embedded in Tissue-Tek optimal cutting temperature (OCT) compound and frozen in liquid nitrogen. Cryostat sections 7 to 8 μm thick were mounted on silane-coated glass slides (Matsunami, Osaka, Japan) and air-dried. Then, the tissues were incubated for 2 hours at room temperature with fluorescein-labeled lotus tetragonolobus lectin (a proximal tubule marker) (Vector Labs, Burlingame, CA). Finally, fluorescence was detected by observation under a fluorescence microscope (models BH2-RFL-T3 and BX50, Olympus, Tokyo, Japan) and processed with Adobe Photoshop software (Adobe Systems, San Jose, CA).
Immunofluorescent microscopy
To evaluate the cells that were positive for Smad4 in each group, paraffin-embedded sections of kidneys were deparaffinized, rehydrated and autoclaved for 15 min at 121°C in 10 mM citrate buffer (pH 6.0) to retrieve antigens, followed by incubation with 10% normal goat serum in PBS to block nonspecific binding of antibodies. Then, the sections of pretreated kidneys were incubated overnight at 4°C with a polyclonal antibody against Smad4 (Abcam, Cambridge, England) at a dilution of 1:200. Subsequently, after washing with PBS, sections were incubated for 2 hours at room temperature with a monoclonal antibody against anti-rabbit IgG conjugated to Alexa 488 (Jackson ImmunoResearch Laboratories, Inc., PA, USA) at a dilution of 1:200. To evaluate the cells that were positive for α-SMA in each group, paraffin-embedded sections of kidneys were incubated overnight at 4°C with a monoclonal antibody against α-SMA conjugated to Cy3 (Sigma Chemical Co.) at a dilution of 1:400 after retrieving antigens and blocking. Then, all of the sections were mounted with the VECTASHIELD mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories Inc., Burlingame, CA, USA) to stain the nucleus. Subsequently, fluorescence was detected by observation under a fluorescence microscope (models BH2-RFL-T3 and BX50, Olympus, Tokyo, Japan) and processed with Adobe Photoshop software (Adobe Systems, San Jose, CA).
Laboratory methods
Blood analysis was performed by a clinical chemistry laboratory (SRL, Tokyo, Japan).
Statistical analysis
The data are expressed as the mean ± standard error (SE). Analysis of variance (ANOVA) was employed to investigate differences among the groups. If statistical significance was detected by ANOVA, Tukey's test was performed as post hoc analysis to compare the means of two different groups. SPSS Statistics version 21 software (IBM, Armonk, NY, USA) was used for statistical analyses. Values of p < 0.05 were considered to be significant.
References
Nath, K. A. Tubulointerstitial changes as a major determinant in the progression of renal damage. Am J Kidney Dis 20, 1–17 (1992).
Becker, G. J. & Hewitson, T. D. The role of tubulointerstitial injury in chronic renal failure. Curr Opin Nephrol Hypertens 9, 133–138 (2000).
Remuzzi, G. & Bertani, T. Pathophysiology of progressive nephropathies. N Engl J Med 339, 1448–1456 (1998).
Campanholle, G., Ligresti, G., Gharib, S. A. & Duffield, J. S. Cellular mechanisms of tissue fibrosis. 3. Novel mechanisms of kidney fibrosis. Am J Physiol Cell Physiol 304, C591–603 (2013).
Humphreys, B. D. et al. Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. Am J Pathol 176, 85–97 (2010).
Lin, S. L., Kisseleva, T., Brenner, D. A. & Duffield, J. S. Pericytes and perivascular fibroblasts are the primary source of collagen-producing cells in obstructive fibrosis of the kidney. Am J Pathol 173, 1617–1627 (2008).
Iwano, M. et al. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J Clin Invest 110, 341–350 (2002).
Kalluri, R. & Neilson, E. G. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest 112, 1776–1784 (2003).
Li, J., Qu, X. & Bertram, J. F. Endothelial-myofibroblast transition contributes to the early development of diabetic renal interstitial fibrosis in streptozotocin-induced diabetic mice. Am J Pathol 175, 1380–1388 (2009).
Broekema, M. et al. Bone marrow-derived myofibroblasts contribute to the renal interstitial myofibroblast population and produce procollagen I after ischemia/reperfusion in rats. J Am Soc Nephrol 18, 165–175 (2007).
Li, J., Deane, J. A., Campanale, N. V., Bertram, J. F. & Ricardo, S. D. The contribution of bone marrow-derived cells to the development of renal interstitial fibrosis. Stem Cells 25, 697–706 (2007).
LeBleu, V. S. et al. Origin and function of myofibroblasts in kidney fibrosis. Nat Med 19, 1047–1053 (2013).
Hannon, G. J. RNA interference. Nature 418, 244–251 (2002).
Tijsterman, M., Ketting, R. F. & Plasterk, R. H. The genetics of RNA silencing. Annu Rev Genet 36, 489–519 (2002).
Dykxhoorn, D. M. & Lieberman, J. The silent revolution: RNA interference as basic biology, research tool and therapeutic. Annu Rev Med 56, 401–423 (2005).
Sledz, C. A. & Williams, B. R. RNA interference in biology and disease. Blood 106, 787–794 (2005).
Leung, R. K. & Whittaker, P. A. RNA interference: from gene silencing to gene-specific therapeutics. Pharmacol Ther 107, 222–239 (2005).
de Fougerolles, A., Vornlocher, H. P., Maraganore, J. & Lieberman, J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov 6, 443–453 (2007).
Takabatake, Y. et al. Chemically modified siRNA prolonged RNA interference in renal disease. Biochem Biophys Res Commun 363, 432–437 (2007).
Shimizu, H. et al. siRNA-based therapy ameliorates glomerulonephritis. J Am Soc Nephrol 21, 622–633 (2010).
Luo, G. H. et al. Inhibition of connective tissue growth factor by small interfering RNA prevents renal fibrosis in rats undergoing chronic allograft nephropathy. Transplant Proc 40, 2365–2369 (2008).
Molitoris, B. A. et al. siRNA targeted to p53 attenuates ischemic and cisplatin-induced acute kidney injury. J Am Soc Nephrol 20, 1754–1764 (2009).
Eddy, A. A. Molecular basis of renal fibrosis. Pediatr Nephrol 15, 290–301 (2000).
Boor, P. & Floege, J. Chronic kidney disease growth factors in renal fibrosis. Clin Exp Pharmacol Physiol 38, 441–450 (2011).
Hamar, P. et al. Small interfering RNA targeting Fas protects mice against renal ischemia-reperfusion injury. Proc Natl Acad Sci U S A 101, 14883–14888 (2004).
Ma, D. et al. Xenon preconditioning protects against renal ischemic-reperfusion injury via HIF-1alpha activation. J Am Soc Nephrol 20, 713–720 (2009).
Lewis, D. L. & Wolff, J. A. Systemic siRNA delivery via hydrodynamic intravascular injection. Adv Drug Deliv Rev 59, 115–123 (2007).
Larson, S. D., Jackson, L. N., Chen, L. A., Rychahou, P. G. & Evers, B. M. Effectiveness of siRNA uptake in target tissues by various delivery methods. Surgery 142, 262–269 (2007).
van de Water, F. M. et al. Intravenously administered short interfering RNA accumulates in the kidney and selectively suppresses gene function in renal proximal tubules. Drug Metab Dispos 34, 1393–1397 (2006).
Meng, X. M. et al. Disruption of Smad4 impairs TGF-beta/Smad3 and Smad7 transcriptional regulation during renal inflammation and fibrosis in vivo and in vitro. Kidney Int 81, 266–279 (2012).
Ortega, A. et al. Role of parathyroid hormone-related protein in tubulointerstitial apoptosis and fibrosis after folic acid-induced nephrotoxicity. J Am Soc Nephrol 17, 1594–1603 (2006).
Acknowledgements
This work was supported by JSPS KAKENHI Grant Numbers 23659026, a grant from The Kidney Foundation, Japan (JFK 11-3) and Jichi Medical University Young Investigator award grant.
Author information
Authors and Affiliations
Contributions
Y.M. conducted the design of the study and coordination, performed the statistical analysis and drafted the manuscript. H.Y. performed all immunofluorescence analysis. M.W. performed all siRNA injections and blood and tissue sampling. K.I. and S.M. participated in the study design. E.K. and D.N. conceived of the study and participated in its design and coordination.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Morishita, Y., Yoshizawa, H., Watanabe, M. et al. siRNAs targeted to Smad4 prevent renal fibrosis in vivo. Sci Rep 4, 6424 (2014). https://doi.org/10.1038/srep06424
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep06424
- Springer Nature Limited
This article is cited by
-
Therapeutic potential of melatonin in targeting molecular pathways of organ fibrosis
Pharmacological Reports (2024)
-
Where should siRNAs go: applicable organs for siRNA drugs
Experimental & Molecular Medicine (2023)
-
The potential of RNA-based therapy for kidney diseases
Pediatric Nephrology (2023)
-
EGR1 and RXRA transcription factors link TGF-β pathway and CCL2 expression in triple negative breast cancer cells
Scientific Reports (2021)
-
Improving Molecular Therapy in the Kidney
Molecular Diagnosis & Therapy (2020)