Abstract
The relationship between neuron morphology and function is a perennial issue in neuroscience. Information about synaptic integration, network connectivity, and the specific roles of neuronal subpopulations can be obtained through morphological analysis of key neurons within a microcircuit. Here we present morphologies of two classes of brainstem respiratory neurons. First, interneurons derived from Dbx1-expressing precursors (Dbx1 neurons) in the preBötzinger complex (preBötC) of the ventral medulla that generate the rhythm for inspiratory breathing movements. Second, Dbx1 neurons of the intermediate reticular formation that influence the motor pattern of pharyngeal and lingual movements during the inspiratory phase of the breathing cycle. We describe the image acquisition and subsequent digitization of morphologies of respiratory Dbx1 neurons from the preBötC and the intermediate reticular formation that were first recorded in vitro. These data can be analyzed comparatively to examine how morphology influences the roles of Dbx1 preBötC and Dbx1 reticular interneurons in respiration and can also be utilized to create morphologically accurate compartmental models for simulation and modeling of respiratory circuits.
Design Type(s) | organism part comparison design |
Measurement Type(s) | neuronal morphology phenotype |
Technology Type(s) | confocal microscopy |
Factor Type(s) | genotype • cell • tissue |
Sample Characteristic(s) | Mus musculus • pre-Botzinger complex • reticular formation |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Similar content being viewed by others
Background & Summary
Neuronal morphology, particularly the structure of the dendritic tree, influences how a neuron integrates synaptic inputs and generates physiological output patterns. Axon projections provide information about connectivity patterns in microcircuits. This study documents the morphology of brainstem interneurons that generate and control breathing.
Breathing is a rhythmic motor behavior that ventilates the lungs to support respiration and homeostasis in air-breathing vertebrates. For humans and all mammals, rodents serve as an advantageous model systesm to study the neural origins of breathing. Key interneuron populations that generate inspiratory, expiratory, and (very recently) post-inspiratory related rhythms have been characterized in terms of physiology, genetic background, and transmitter phenotype1,2,3,4. Premotor neurons that influence airway resistance have been similarly characterized5,6,7,8,9,10,11. However, only a limited number of morphologies of constituent neurons in these populations have been documented and analyzed5,12,13. This data descriptor aims to ameliorate that problem by providing annotated, high-quality digital reconstructions of the morphologies of rhythm-generating interneurons and motor pattern-related premotor neurons from neonatal mice.
The respiratory cycle is dominated by the rhythm underlying inspiration, which is generated within the preBötzinger complex (preBötC) of the ventral medulla1,2,3,4,14,15. Rhythmogenic preBötC neurons are derived from precursor cells that express the homeobox transcription factor Dbx1 (refs 13, 16,17,18), hereafter referred to as Dbx1 neurons. The intermediate reticular formation, immediately adjacent (dorsal) to the preBötC, is a diverse region containing respiratory Dbx1 premotor interneurons that control inspiratory related muscles of the tongue and pharynx9,19,20,21.
In this study we used intersectional mouse genetics to induce fluorescent protein expression in Dbx1 neurons of neonatal mice. Neuronal morphologies were acquired following patch-clamp recordings in transverse brainstem slices that retain the preBötC, the intermediate reticular formation, as well as the hypoglossal (XII) motor nucleus. These slices expose the preBötC and reticular formation at the rostral surface and spontaneously generate inspiratory rhythm and XII motor output, thus providing an experimentally advantageous breathing model in vitro22,23.
We obtained three-dimensional morphologies of respiratory Dbx1 preBötC and Dbx1 intermediate reticular formation neurons by filling neurons with biocytin during whole-cell patch-clamp recordings24,25,26,27. Compared to other reconstruction methods such as fluorescence microscopy of dye-filled neurons, biocytin reconstructions can be more time consuming but provide better visualization of thinner neuronal processes and axons28. Once labeled, we visualized the recorded neurons via confocal imaging and manually reconstructed their morphologies in a convenient digital format suitable for storage, display, and analysis.
Over the past four years, our laboratory contributed 47 digital neuronal morphologies to the public open access database NeuroMorpho.org. Of those 47 digital reconstructions, 23 correspond to Dbx1 preBötC neurons12,13 (six have not been previously published; this report describes them for the first time). Twelve of the 47 correspond to preBötC neurons not derived from Dbx1-expressing precursors12 (i.e., non-Dbx1 preBötC neurons), and 12 correspond to Dbx1 reticular formation neurons5.
Digital morphologies can be analyzed by software packages such as L-measure29, which computes more than 40 different morphometric properties of dendritic trees and axons. Sholl analysis, which provides branching and dendritic density information in regular distance intervals from the soma30,31, can be performed with software such as NeuronStudio32. Digital morphologies can also be readily ported to simulation packages such as NEURON33 and GENESIS34 to form compartmental mathematical models that are high-fidelity representations of real neurons. We intend that these morphological data be meta-analyzed and incorporated into models of inspiratory rhythm- and pattern-generating circuits of the lower brainstem to better understand the neural mechanisms of breathing.
Methods
Mice
All of the animal protocols were approved by the Institutional Animal Care and Use Committee at The College of William and Mary, which follows the guidelines provided by the US National Institutes of Health Office of Laboratory Animal Welfare35.
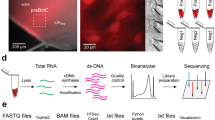
Figure 1 recaps the workflow, which is detailed below. We crossed female mice that express Cre recombinase fused to a tamoxifen-sensitive estrogen receptor (CreERT2) under the control of the Dbx1 promoter, i.e., Dbx1CreERT2 (stock no. 028131, Jackson labs, Bar Harbor, ME)36 with floxed male reporter mice that express red fluorescent protein variant tdTomato in a Cre-dependent manner (Rosa26tdTomato, stock no. 007905, Jackson labs)37. Offspring with both alleles (Dbx1CreERT2; Rosa26tdTomato mice), whose pregnant dams received tamoxifen during embryonic development, express the fluorescent reporter in Dbx1-derived cells12,22,36 (Fig. 1, step 1). Dbx1CreERT2 mice were maintained on a CD-1 background strain. Rosa26tdTomato reporter mice were maintained using a C57BL/6J background strain.
Expression of the red fluorescent protein tdTomato is induced in Dbx1-derived neurons (and glia) in mice using intersectional mouse genetic technologies (1). A transverse slice of the brainstem (indicated by the gray box) containing the preBötzinger complex is taken from a neonatal transgenic mouse (2). The slice is used for physiology recordings, during which respiratory modulated neurons are filled with biocytin (3). The slice is then preserved in 4% paraformaldehyde (4) and made transparent via incubation in Scale solution (5). The slice is treated with ExtrAvidin FITC (6) which binds to the biocytin allowing for visualization of the neuron through confocal microscopy (7). Confocal images in the x-, y-, and z- dimensions are taken of the entire neuron morphology and stitched together using FIJI (8). Using the 3D confocal images, neurons are digitally reconstructed using Neuromantic (9). XII, hypoglossal nucleus; NA, nucleus ambiguus, preBötC, preBötzinger complex; IO, inferior olive.
Dbx1CreERT2 mice were also mated with floxed reporter mice that express a channelrhodopsin-2/tdTomato fusion protein (Rosa26ChR2-tdTomato, stock no. 12567, Jackson labs)38. The Dbx1CreERT2; Rosa26ChR2-tdTomato mice were employed in separate electrophysiological experiments; here we recovered the morphology of the recorded neurons in the same way as Dbx1CreERT2; Rosa26tdTomato, which was possible because both expressed native tdTomato in Dbx1-derived neurons. Channelrhodopsin, while important for physiological tests, has no impact on morphological studies. Figure 1 only indicates Dbx1CreERT2; Rosa26tdTomato mice for simplicity.
Animal genotypes were verified using real-time PCR using primers for Cre and tandem dimer red fluorescent protein (Transnetyx, Cordova, TN). Timed matings were monitored such that embryonic day 0.5 (E0.5) was defined as 12 h after the start of cohabitation. Cre recombination was then induced by administering tamoxifen (T5648; Sigma Aldrich, St Louis, MO) at E10.5 when Dbx1 is at or near peak expression in the hindbrain16,17,36,39. Tamoxifen was administered by oral gavage to pregnant dams at a concentration of 0.9 mg/40 g body mass.
Transverse slice preparations
Neonatal Dbx1CreERT2; Rosa26tdTomato and Dbx1CreERT2; Rosa26ChR2-tdTomato mice were anesthetized then euthanized via decapitation at postnatal days 0–5 (P0–5), consistent with protocols outlined by the American Veterinary Medical Association Guidelines for euthanasia of animals40. Transections were made at the bregma and the thorax. The neuraxis, from the pons to the lower thoracic spinal cord, was then removed within two minutes and further dissected in artificial cerebrospinal fluid (ACSF) containing (mM): 124 NaCl, 3 KCl, 1.5 CaCl2, 1 MgSO4, 25 NaHCO3, 0.5 NaH2PO4 and 30 dextrose, equilibrated with 95% O2 and 5% CO2 (pH 7.4) (Fig. 1, step 2). The neuraxis was then glued to an agar block with the ventral surface facing out and placed in the vise of a vibratome. We cut 550-μm-thick transverse brainstem slices that exposed the preBötC at the rostral face and retained the rostral XII nerve rootlets22 (Fig. 1, step 3). Slices were perfused with ACSF at 28 °C in a recording chamber on a fixed-stage upright microscope equipped with differential interference contrast optics and epifluorescence, which enables visual identification and selective recording of target neurons. The K+ concentration in the ACSF was elevated to 9 mM to maintain long-term stability of the preBötC rhythm22,23,41. Rhythmic inspiratory-related motor output was recorded from the XII nerve rootlets using suction electrodes and a differential amplifier. Whole-cell patch-clamp recordings were acquired using capillary glass micro-pipettes and a current-clamp amplifier. Patch pipettes were positioned under visual control after fluorescent identification of Dbx1 neurons. The patch solution contained (mM): 140 potassium gluconate, 10 Hepes, 5 NaCl, 1 MgCl2, 0.1 EGTA, 2 Mg-ATP, 0.3 Na3-GTP and 2 mgml−1 biocytin (B4261; Sigma Aldrich). All of the neurons in this data set were rhythmically active in sync with inspiratory XII motor output.
After the recordings, transverse slices containing biocytin-filled neurons were fixed in 4% paraformaldehyde in 0.1 M sodium phosphate buffer for at least 16 h at 4 °C (Fig. 1, step 4). Then, the slices were treated with Scale solution containing 4 M urea, 10% (mass/volume) glycerol and 0.1% (m/v) Triton X-100, for 10 days to clear the tissue and remove opaque background staining42 (Fig. 1, step 5). Slices were washed three times for 15 min each in phosphate buffer solution (PBS)+1% Triton X-100 (PBST) and then blocked in PBST with 10% heat-inactivated fetal bovine sera (F4135; Sigma Aldrich) for 45 min. The biocytin was revealed by incubating the slices with fluorescein isothiocyanate-conjugated ExtrAvidin (E2761; Sigma Aldrich) overnight at 4 °C with three-dimensional rotation on a nutator (Fig. 1, step 6). Next, the slices were rinsed with PBS five times for 15 min each and cover-slipped in Vectashield (H-1500; Vector Laboratories, Burlingame, CA).
Confocal microscopy and digital neuronal reconstruction
We visualized recorded neurons using a spinning-disk confocal microscope (Olympus BX51, Center Valley, PA) and a laser scanning confocal microscope (Zeiss LSM 510, Thornwood, NY) Three-dimensional (3D) confocal images of the individual neurons were obtained using a 20x objective (Olympus numerical aperture 0.5, Zeiss LSM numerical aperture 1.0) at increments of 1 μm in the z-axis (Fig. 1, step 7). The series of confocal images (i.e., z-stacks) were aligned in three-dimensions, merged or ‘stitched together’ at contiguous borders using ImageJ software43 and the Stitching plugin44 (Fig. 1, step 8). This stitching process was iterated until the entire morphology of the neuron was contained within a single three-dimensional image file. Finally we digitized neuronal morphologies using the Neuromantic reconstruction tool, which is also free and in the public domain45. The digital reconstructions were scaled to the appropriate size based on the micron-to-pixel ratio for each microscope (Fig. 1, step 9). Images acquired from the LSM microscope were scaled with a 0.41 micron-to-pixel ratio and images from the Olympus microscope were scaled using a 0.322 micron-to-pixel ratio. This data descriptor pertains to 47 digital morphologies of inspiratory modulated Dbx1 preBötC neurons, six of which are previously unpublished (Data Citations 1–6) and 41 which are associated with previous publications (Data Citations 7–47). The morphologies are all publicly available via NeuroMorpho.org.
Data Records
Digital reconstructions of Dbx1 neurons are located in the Del Negro archive of the NeuroMorpho database (Data Citations 1–47). Digital reconstruction files are in SWC format, which is a commonly used format for neuron morphologies24. The reconstruction files contain an x-coordinate, y-coordinate, and z-coordinate of each neuronal segment. The type of neuronal process, such as cell body, axon, or dendrite is also specified by type 1, 2, and 4, respectively. (Type 3 represents basal dendrites, but there is no such distinction in brainstem interneurons, so type 3 is omitted as a classifier in our dataset. Our dendrites were all designated type 4.) The radius in microns is given for each neuronal segment as well as the ‘parent’ segment or the index number of the previous segment. Table 1 provides an example of an SWC file output for a neuron reconstruction. Physiological properties of Dbx1 preBötC and Dbx1 reticular neurons have been described5,12,13. Table 2 lists the reconstructions available in the Del Negro archive of NeuroMorpho.org.
Technical Validation
In newborn Dbx1CreERT2; Rosa26tdTomato and Dbx1CreERT2; Rosa26ChR2-tdTomato mice, Dbx1 neurons form an inverted U-shape in the transverse (coronal) plane, which is visible in brainstem slices at the level of the preBötC. The inverted U-shape originates at the lateral border of the hypoglossal motor nucleus, located within the dorso-medial portion of the slice, and continues ventrolaterally until the ventral border of the tissue slice22. The dorsal border of the preBötC is identifiable because it is immediately ventral to the semi-compact division of the nucleus ambiguus, which does not express Dbx122. Visual identification of the principal loop of the inferior olive and the flattening of the V-shape of the fourth ventricle are other indicators that the rostral surface of the transverse slice is at the level of the preBötC22.
Slices remained in the recording chamber for at least 15 min after biocytin dialysis to maximize biocytin diffusion throughout the cytoplasm12,24. A clearing agent was used to facilitate visualization of the morphology; however clearing reagents can cause tissue shrinkage or expansion which could distort morphological features42. The Scale solution used to clear the tissue in these experiments minimizes or completely precludes tissue expansion (compared to other methods)42.
The quality of digital reconstructions depends on histology methods, image acquisition, as well as the digital reconstruction algorithms. To minimize disparities, we consistently used the same method of histological labeling. The software Neuromantic45, used for digitizing our image stacks, offers up to 16,000% magnification. This zoom feature enables the user to adhere to the most minute details captured in the image, which results in the most accurate reconstruction possible.
Two of the six new neurons and nine of the previously published neurons had no discernible axon, which might have indicated insufficient biocytin filling or that the axon was severed during tissue preparation. We recommend that the end user of the data draw no firm conclusions regarding connectivity from the lack of an axon in reconstructed digital morphology.
For those neurons whose axons were discernible, we distinguished the axons from the dendrites according to these criteria: 1) axons generally have a constant diameter whereas dendrites taper distal to the soma; 2) axons exhibit fewer branches and never show spine-like protrusions; 3) truncated axons near the slice surface exhibit a bleb or fluorescent circle from the cut end46,47.
Digital reconstructions were uploaded to NeuroMorpho.org, where they undergo a standardization process. The soma (type 1) should be the initial parent segment for all subsequent segments, whether dendritic or axonal. Neuronal processes should only connect to either the soma or to segments of the same type; for example, dendrite segments connect to dendrite segments and axon segments connect to axon segments. All processes should have a designated type and should not be undefined. A process can branch into no more than two processes at any given point.
Some irregularities can be fixed automatically during the standardization process48. If a neuronal segment is designated as a different type than its parent and daughter segments (e.g., a type 3 surrounded by type 2 s) the erroneous segment type is automatically changed to match the type of its parent and daughter segments. If the soma is not the initial segment in the file, the soma segment is automatically changed to the first segment in the file. If a segment has a radius of zero microns, then the radius is automatically changed to match the radius of its parent. Other digitization issues must be corrected by the submitting investigator48. For example, if a segment has not been designated with a process type, the correct type must be manually entered, rather than automatically assigned, which ensures that the proper type has been documented. Segments with a radius of zero (i.e., less than 0.05 μm), or larger than four standard deviations above the average radius of the cell are flagged as physiologically unrealistic during standardization and must be resolved by the submitting investigator. After the standardization process, digital reconstruction files and images are then reviewed and approved by the submitting investigator before being added to the public database48.
Additional Information
How to cite this article: Akins, V. T. et al. Morphology of Dbx1 respiratory neurons in the preBötzinger complex and reticular formation of neonatal mice. Sci. Data 4:170097 doi: 10.1038/sdata.2017.97 (2017).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
References
Feldman, J. L. & Del Negro, C. A. Looking for inspiration: new perspectives on respiratory rhythm. Nat. Rev. Neurosci. 7, 232–241 (2006).
Feldman, J. L., Del Negro, C. A. & Gray, P. A. Understanding the rhythm of breathing: so near, yet so far. Annu. Rev. Physiol. 75, 423–452 (2013).
Richter, D. W. & Smith, J. C. Respiratory rhythm generation in vivo. Physiol. Bethesda Md 29, 58–71 (2014).
Ramirez, J.-M., Dashevskiy, T., Marlin, I. A. & Baertsch, N. Microcircuits in respiratory rhythm generation: commonalities with other rhythm generating networks and evolutionary perspectives. Curr. Opin. Neurobiol. 41, 53–61 (2016).
Revill, A. L. et al. Dbx1 precursor cells are a source of inspiratory XII premotoneurons. eLife 4, e12301 (2015).
Ono, T., Ishiwata, Y., Inaba, N., Kuroda, T. & Nakamura, Y. Modulation of the inspiratory-related activity of hypoglossal premotor neurons during ingestion and rejection in the decerebrate cat. J. Neurophysiol. 80, 48–58 (1998).
Gestreau, C., Dutschmann, M., Obled, S. & Bianchi, A. L. Activation of XII motoneurons and premotor neurons during various oropharyngeal behaviors. Respir. Physiol. Neurobiol. 147, 159–176 (2005).
Travers, J. B., Yoo, J.-E., Chandran, R., Herman, K. & Travers, S. P. Neurotransmitter phenotypes of intermediate zone reticular formation projections to the motor trigeminal and hypoglossal nuclei in the rat. J. Comp. Neurol. 488, 28–47 (2005).
Chamberlin, N. L., Eikermann, M., Fassbender, P., White, D. P. & Malhotra, A. Genioglossus premotoneurons and the negative pressure reflex in rats. J. Physiol. 579, 515–526 (2007).
Koizumi, H. et al. Functional imaging, spatial reconstruction, and biophysical analysis of a respiratory motor circuit isolated in vitro. J. Neurosci. 28, 2353–2365 (2008).
Volgin, D. V., Rukhadze, I. & Kubin, L. Hypoglossal premotor neurons of the intermediate medullary reticular region express cholinergic markers. J. Appl. Physiol. Bethesda Md 1985 105, 1576–1584 (2008).
Picardo, M. C. D., Weragalaarachchi, K. T. H., Akins, V. T. & Del Negro, C. A. Physiological and morphological properties of Dbx1 -derived respiratory neurons in the preBötzinger complex of neonatal mice. J. Physiol. 591, 2687–2703 (2013).
Wang, X. et al. Laser ablation of Dbx1 neurons in the pre-Bötzinger complex stops inspiratory rhythm and impairs output in neonatal mice. eLife 3, e03427 (2014).
Smith, J. C., Ellenberger, H. H., Ballanyi, K., Richter, D. W. & Feldman, J. L. Pre-Bötzinger complex: a brainstem region that may generate respiratory rhythm in mammals. Science 254, 726–729 (1991).
Moore, J. D. et al. Hierarchy of orofacial rhythms revealed through whisking and breathing. Nature 497, 205–210 (2013).
Bouvier, J. et al. Hindbrain interneurons and axon guidance signaling critical for breathing. Nat. Neurosci. 13, 1066–1074 (2010).
Gray, P. A. et al. Developmental origin of preBötzinger complex respiratory neurons. J. Neurosci. 30, 14883–14895 (2010).
Vann, N. C., Pham, F. D., Hayes, J. A., Kottick, A. & Negro, C. A. D. Transient suppression of Dbx1 preBötzinger interneurons disrupts breathing in adult mice. PLOS ONE 11, e0162418 (2016).
Peever, J. H., Shen, L. & Duffin, J. Respiratory pre-motor control of hypoglossal motoneurons in the rat. Neuroscience 110, 711–722 (2002).
Stanek, E., Cheng, S., Takatoh, J., Han, B.-X. & Wang, F. Monosynaptic premotor circuit tracing reveals neural substrates for oro-motor coordination. eLife 3, e02511 (2014).
Welzl, H. & Bureš, J. Lick-synchronized breathing in rats. Physiol. Behav. 18, 751–753 (1977).
Ruangkittisakul, A., Kottick, A., Picardo, M. C. D., Ballanyi, K. & Negro, C. A. D. Identification of the pre-Bötzinger complex inspiratory center in calibrated ‘sandwich’ slices from newborn mice with fluorescent Dbx1 interneurons. Physiol. Rep. 2, e12111 (2014).
Funk, G. D. & Greer, J. J. The rhythmic, transverse medullary slice preparation in respiratory neurobiology: contributions and caveats. Respir. Physiol. Neurobiol. 186, 236–253 (2013).
Jacobs, G., Claiborne, B. & Harris, K. in Computational Modeling Methods for Neuroscientists (ed. de Schutter, E. 187–210 (MIT Press, 2010).
Halavi, M., Hamilton, K. A., Parekh, R. & Ascoli, G. A. Digital reconstructions of neuronal morphology: three decades of research trends. Front. Neurosci 6, 49 (2012).
Parekh, R. & Ascoli, G. A. Neuronal morphology goes digital: a research hub for cellular and system neuroscience. Neuron 77, 1017–1038 (2013).
Parekh, R. & Ascoli, G. A. Quantitative investigations of axonal and dendritic arbors: development, structure, function, and pathology. The Neuroscientist 21, 241–254 (2015).
Blackman, A. V., Grabuschnig, S., Legenstein, R. & Sjöström, P. J. A comparison of manual neuronal reconstruction from biocytin histology or 2-photon imaging: morphometry and computer modeling. Front. Neuroanat 8, 65 (2014).
Scorcioni, R., Polavaram, S. & Ascoli, G. A. L-Measure: a web-accessible tool for the analysis, comparison and search of digital reconstructions of neuronal morphologies. Nat. Protoc. 3, 866–876 (2008).
Sholl, D. A. Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat. 87, 387–406 (1953).
Langhammer, C. G. et al. Automated Sholl analysis of digitized neuronal morphology at multiple scales: whole cell Sholl analysis versus Sholl analysis of arbor subregions. Cytometry A 77A, 1160–1168 (2010).
Wearne, S. L. et al. New techniques for imaging, digitization and analysis of three-dimensional neural morphology on multiple scales. Neuroscience 136, 661–680 (2005).
Hines, M. L. & Carnevale, N. T. The NEURON simulation environment. Neural Comput. 9, 1179–1209 (1997).
Bower, J. M., Beeman, D. & Hucka, M. in The Handbook of Brain Theory and Neural Networks (ed. Arbib, M. A. 475–478 (MIT Press, 2003).
NIH Office of Laboratory Animal Welfare. Public Health Service Policy on Humane Care and Use of Laboratory Animals (Office of Laboratory Animal Welfare, National Institutes of Health, Department of Health and Human Services, 2015).
Hirata, T. et al. Identification of distinct telencephalic progenitor pools for neuronal diversity in the amygdala. Nat. Neurosci. 12, 141–149 (2009).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Madisen, L. et al. A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat. Neurosci. 15, 793–802 (2012).
Pierani, A. et al. Control of interneuron fate in the developing spinal cord by the progenitor homeodomain protein Dbx1. Neuron 29, 367–384 (2001).
Leary, S. et al. AVMA Guidelines for the Euthanasia of Animals: 2013 edition (American Veterinary Medical Association, 2013).
Ruangkittisakul, A. et al. High sensitivity to neuromodulator-activated signaling pathways at physiological [K+] of confocally imaged respiratory center neurons in on-line-calibrated newborn rat brainstem slices. J. Neurosci. 26, 11870–11880 (2006).
Hama, H. et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 14, 1481–1488 (2011).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Preibisch, S., Saalfeld, S. & Tomancak, P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25, 1463–1465 (2009).
Myatt, D. R., Hadlington, T., Ascoli, G. A. & Nasuto, S. J. Neuromantic—from semi-manual to semi-automatic reconstruction of neuron morphology. Front. Neuroinformatics 6, 4 (2012).
Furness, J. B., Alex, G., Clark, M. J. & Lal, V. V. Morphologies and projections of defined classes of neurons in the submucosa of the guinea-pig small intestine. Anat. Rec. A. Discov. Mol. Cell. Evol. Biol 272A, 475–483 (2003).
Swietek, B., Gupta, A., Proddutur, A. & Santhakumar, V. Immunostaining of biocytin-filled and processed sections for neurochemical markers. J. Vis. Exp. 118, e54880 (2016).
Halavi, M. et al. NeuroMorpho.org implementation of digital neuroscience: dense coverage and integration with the NIF. Neuroinformatics 6, 241 (2008).
Data Citations
Del Negro, C. A. NeuroMorpho.org NMO_45917 (2017)
Del Negro, C. A. NeuroMorpho.org NMO_45918 (2017)
Del Negro, C. A. NeuroMorpho.org NMO_45921 (2017)
Del Negro, C. A. NeuroMorpho.org NMO_45922 (2017)
Del Negro, C. A. NeuroMorpho.org NMO_45923 (2017)
Del Negro, C. A. NeuroMorpho.org NMO_45924 (2017)
Del Negro, C. A. NeuroMorpho.org NMO_09581 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09582 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09583 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09584 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09585 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09586 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09587 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09588 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09589 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09590 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09591 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09592 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09592 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09594 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09595 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09596 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09597 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09598 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09599 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09600 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09601 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09602 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09603 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09604 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09605 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_09606 (2013)
Del Negro, C. A. NeuroMorpho.org NMO_45919 (2016)
Del Negro, C. A. NeuroMorpho.org NMO_45920 (2016)
Del Negro, C. A. NeuroMorpho.org NMO_45925 (2016)
Del Negro, C. A. NeuroMorpho.org NMO_45926 (2016)
Del Negro, C. A. NeuroMorpho.org NMO_45927 (2016)
Del Negro, C. A. NeuroMorpho.org NMO_45928 (2016)
Del Negro, C. A. NeuroMorpho.org NMO_45929 (2016)
Del Negro, C. A. NeuroMorpho.org NMO_45930 (2016)
Del Negro, C. A. NeuroMorpho.org NMO_45931 (2016)
Del Negro, C. A. NeuroMorpho.org NMO_45932 (2016)
Del Negro, C. A. NeuroMorpho.org NMO_45933 (2016)
Del Negro, C. A. NeuroMorpho.org NMO_45934 (2016)
Del Negro, C. A. NeuroMorpho.org NMO_45935 (2016)
Del Negro, C. A. NeuroMorpho.org NMO_45936 (2016)
Del Negro, C. A. NeuroMorpho.org NMO_45937 (2016)
Acknowledgements
The authors thank Araya Ruangkittisakul and Andrew Kottick for filling neurons with biocytin that became part of this data set. This work was supported by grants from the National Institutes of Health to PI C.A.D.N. (R01 HL-104127, R21 NS-070056, and R21 NS-087257).
Author information
Authors and Affiliations
Contributions
V.T.A. and C.A.D.N. drafted the manuscript. V.T.A. performed the histology, acquired morphologies, and performed morphometric analysis of Dbx1 neurons in preBötC and reticular formation. K.W. acquired morphologies and performed morphometric analysis of Dbx1 and non-Dbx1 neurons. A.L.R. and M.C.D.P. patch-recorded the neurons and filled them with biocytin.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
ISA-Tab metadata
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/ The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files made available in this article.
About this article
Cite this article
Akins, V., Weragalaarachchi, K., Picardo, M. et al. Morphology of Dbx1 respiratory neurons in the preBötzinger complex and reticular formation of neonatal mice. Sci Data 4, 170097 (2017). https://doi.org/10.1038/sdata.2017.97
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/sdata.2017.97
- Springer Nature Limited
This article is cited by
-
Fine mapping and candidate gene analysis of Dravet syndrome modifier loci on mouse chromosomes 7 and 8
Mammalian Genome (2024)
-
Caffeine exacerbates seizure-induced death via postictal hypoxia
Scientific Reports (2023)
-
An open repository for single-cell reconstructions of the brain forest
Scientific Data (2018)