Abstract
E2F3 and MYC are transcription factors that control cellular proliferation. To study their mechanism of action in the context of a regenerating tissue, we isolated both proliferating (crypts) and non-dividing (villi) cells from wild-type and Rb depleted small intestines of mice and performed ChIP-exo-seq (chromatin immunoprecipitation combined with lambda exonuclease digestion followed by high-throughput sequencing). The genome-wide chromatin occupancy of E2F3 and MYC was determined by mapping sequence reads to the genome and predicting preferred binding sites (peaks). Binding sites could be accurately identified within small regions of only 24 bp-28 bp long, highlighting the precision to which binding peaks can be identified by ChIP-exo-seq. Forty randomly selected E2F3- and MYC-specific binding sites were validated by ChIP-PCR. In addition, we also presented gene expression data sets from wild type, Rb-, E2f3- and Myc-depleted crypts and villi within this manuscript. These represent comprehensive and validated datasets that can be integrated to identify putative direct targets of E2F3 and MYC involved in the control of cellular proliferation in normal and Rb-deficient small intestines.
Design Type(s) | parallel group design • genetic modification design |
Measurement Type(s) | chromatin binding • transcription profiling assay |
Technology Type(s) | transcription factor binding site identification by ChIP-Seq assay • DNA microarray |
Factor Type(s) | Genetic Variation |
Sample Characteristic(s) | Mus musculus • small intestine |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Similar content being viewed by others
Background & Summary
E2F is a family of transcription factors that links up-stream proliferation signals with the timely expression of many downstream cell cycle regulated genes1. Among the E2F members, E2F3 is known as a major transcriptional activator that can be inhibited when in complex with RB. During late G1 phase of the cell cycle, CDK mediated phosphorylation of RB leads to the release of E2F3 from RB-E2F3 complexes and the activation of the G1–S transcriptional program, resulting in entry of cells into S phase1. MYC is another important transcriptional regulator of the cell cycle2. It’s an oncogene with a broad functional spectrum, which includes the control of cellular proliferation, cell survival, cell growth and metabolism3. Despite significant functional overlap between E2F and MYC in the control of cell cycle regulation, the mechanisms linking these two critical transcriptional programs are poorly understood.
The small intestine has been utilized as a model system to study normal cellular proliferation and cancer. A unique characteristic of the small intestine is that the continuous regeneration of the entire intestinal track is neatly organized in a defined pouch-like invagination called the crypt4. At the base of crypts, pluripotent stem cells asymmetrically divide to generate a new stem cell and a transient amplifying progenitor cell. Progenitor cells undergo multiple rounds of cell division to form a transient amplifying zone. As progenitor cells migrate towards the finger-like protrusions called villi, they exit the cell cycle and differentiate4. Thus, proliferating progenitor cells and differentiated enterocytes are located in distinct spatial regions of the intestinal track as depicted in Fig. 1. We chose to evaluate E2F3 and MYC chromatin occupancy in cells of the small intestine in order to better define how these two factors regulate the cell cycle in vivo.
ChIP-seq is a widely used technique to profile chromatin occupancy of transcription factors on a genome wide scale. Briefly, chromatin is released from lysed cells and sheared to smaller fragments and specific antibodies are used to immunoprecipitate protein-DNA complexes. Then, the enriched DNA fragments are purified, tagged, amplified and sequenced using next-generation sequencing technology. Finally, genomic regions with enriched read alignments are detected as predicted protein binding sites, which usually span hundreds of base pairs5,6. Recently, an improved technique to better define chromatin binding regions was developed, ChIP-exo-seq7,8. The key innovation of this technique is that after ChIP, the lambda exonuclease is used to digest DNA fragments starting from the exposed 5′ end and stopping at the protein-DNA boundary. After library preparation and deep sequencing, the 5′ end of the reads obtained are highly concentrated on the protein-DNA boundary, providing better positional resolution and accuracy of the predicted protein binding sites7,8.
Both ChIP-exo-seq experiments and expression microarray experiments were conducted with enriched crypt and villus fractions obtained from the small intestine of mice (Table 1, available online only). For ChIP-exo-seq experiments, E2F3 and MYC chromatin binding was analyzed in parallel in crypts and villi derived from wild type and Rb deficient small intestines. An average of 70 million to 120 million reads were generated for each ChIP-exo-seq experiment (Table 2). For gene expression assays, we used an Affymetrix microarray platform (Mouse Genome 430 2.0 Array) to profile mRNA levels in crypts and villi derived from various genetically modified mice with respect to E2f, Myc and Rb deficiency (Table 1, available online only). The intersection between ChIP-exo-seq and mRNA expression data sets are identified as putative direct targets of MYC and E2F3. Comparison of putative MYC and E2F3 target genes revealed unique and overlapping sets of targets, suggesting distinct and synergistic roles for MYC and E2F3 in the control of gene expression in the small intestine. Our related work recently published in Nature Cell Biology used these molecular approaches to address different biological questions related to the control of cellular proliferation in vivo9. In this Data Descriptor, we provide additional information aimed to help other investigators interpret and use these data sets for their own research.
Methods
Mouse usage
The description of the mouse strains used and how they were maintained is described in detail in the related research manuscript9. Briefly, mice were housed at the Ohio State University ventilated animal vivarium under standard conditions (temperature at 22 °C and 12 h light/12 h dark cycle per day). Experimental manipulation of animals was approved by Institutional Animal Care and Use Committee at the Ohio State University. Mouse strains were bred and maintained in a mixed genetic background (C57BL/6×129×FVB/N). Both genders of mice were utilized in the studies. To induce the expression of Cre recombinase and deletion of alleles flanked by loxP sequences, β-naphthoflavone (Sigma-Aldrich; N3633) dissolved in corn oil (Sigma-Aldrich; C8267) was intraperitoneally injected into 2-month old mice at the dosage of 80 mg kg−1 body weight10. Five injections were performed within 30 h (9am-3pm-9am on first day and 9am-3pm on second day).
Mouse tissue collection
Immediately after euthanizing mice, the small intestine was dissected and mesentery and adipose tissues were removed. The intestine was then cut open along their cephalocaudal axis and gently washed in phosphate buffered saline (PBS) to remove undigested food chow. The tissue used for RNA isolation was collected from a 10 cm portion of the intestine proximal to the stomach. For ChIP, the intestine was divided into three parts of equal length and the first 1/3 section proximal to the stomach was used. After cleaning with PBS, the tissue was incubated in 25 ml PBS containing 0.5 mM ethylenediaminetetraacetic acid (EDTA) and 1 mM dithiothreitol (DTT). After incubation for 30 min at room temperature, villus fractions were collected by multiple rounds of gently shaking the tissue, decanting tissue materials in suspension into multiple tubes containing 10 ml ice-cold PBS with 1 mM DTT. After most villi were collected, the remaining tissue was incubated in 25 ml PBS with 0.9 mM EDTA and 1 mM DTT. The crypt fractions were collected by shaking the tissue in multiple tubes containing 10 ml ice-cold PBS with 1 mM DTT until complete separation of epithelial cells from mesenchymal tissues. Crypt-enriched fractions were filtered using 70 μm cell strainer (Fisher Scientific; 22363548) to remove potential contamination of broken pieces of villi with larger sizes. After centrifuge at 335×g for 5 min at 4 °C, the resulting villus-enriched or crypt-enriched tissue pellets were combined and washed in ice-cold PBS. The tissues were then precipitated by centrifuge at 335×g for 5 min at 4 °C. For RNA isolation and ChIP assays, the cell pellets were processed as described below.
RNA isolation
Total RNA from purified villi/crypts was isolated using TRIzol reagent following manufacturer’s protocol and further cleaned up with RNeasy Mini Kit (Qiagen; 74104). RNA integrity was assessed using Agilent 2100 Bioanalyzer Instruments. Global RNA expression levels were profiled using Affymetrix GeneChip Mouse Genome 430 2.0 Array at the Ohio State University Shared Resources (http://cancer.osu.edu/research-and-education/shared-resources/genomics/services).
ChIP
Freshly isolated crypts/villi were crosslinked in PBS containing 1% formaldehyde at 37 °C for 15 min on a rotator. The crosslinking reaction was terminated by incubation with 0.125 M glycine at 37 °C for 5 min on a rotator. The cell pellets were then washed in ice-cold PBS, followed by cytosolic lysis and nuclear lysis steps (cytosolic lysis buffer: 5 mM PIPES (pH 8.0), 85 mM KCl, 0.5% NP-40; nuclear lysis buffer: 50 mM Tris (pH 8.0), 10 mM EDTA, 1% SDS). The released chromatin was then subject to sonication to generate DNA fragments mainly with 100-300 bp sizes. The fragmented chromatin was diluted with 9 volumes of IP dilution buffer (16.7 mM Tris (pH 8.0), 167 mM NaCl, 1.2 mM EDTA, 1.1% Triton X-100). To minimize non-specific pull-down, the chromatin was incubated with Protein G Plus/Protein A beads (Calbiochem; IP05) at 4 °C for 1 h on a rotator prior to adding antibodies. Antibodies used in the study were E2F3 (Santa Cruz; sc-878 X), MYC (Santa Cruz; sc-764 X) and normal rabbit IgG (Santa Cruz; sc-2027). The chromatin was incubated with antibodies overnight at 4 °C on a rotator, followed by incubation with Protein G Plus/Protein A beads (Calbiochem; IP05) at 4 °C for 2 h on a rotator. The chromatin-antibody-bead complex was then washed twice with each of the following buffers: low-salt buffer (20 mM Tris (pH 8.0), 2 mM EDTA, 1% Triton X-100, 0.1% SDS and 150 mM NaCl), high-salt buffer (20 mM Tris (pH 8.0), 2 mM EDTA, 1% Triton X-100, 0.1% SDS and 500 mM NaCl), LiCl buffer (10 mM Tris (pH 8.0), 1 mM EDTA, 250 mM LiCl, 1% NP-40 and 1% deoxycholic acid) and Tris-EDTA buffer (pH 8.0). Note that all the steps described above were performed in the presence of proteinase inhibitors. The Protein G Plus/Protein A beads were incubated with 1% SDS plus 0.1M NaHCO3 (900 rpm vortex) to retrieve the immunoprecipitated chromatin. The chromatin was then reverse-crosslinked by incubation with 0.5M NaCl and 0.1 mg/ml RNase A overnight at 65 °C (including non-immunoprecipitated inputs), followed by 0.1 mg/ml proteinase K treatment at 50 °C for 1 h. The DNA was purified with Qiagen kits (QIAquick Purification Kit (Qiagen; 28104). Quantitative PCR was performed using SYBR Green master mix (Bio-Rad; 170-8884) with Applied Biosystems StepOnePlus Real-Time PCR System.
ChIP-exo-seq
For ChIP-exo-seq experiments, crypts were collected and pooled from 32 wild type mice and 27 Rb KO mice and villi were collected from 7 wild type mice and 7 Rb KO mice. The subsequent steps, including crosslinking, cytosolic and nuclear lysis, sonication, dilution with IP dilution buffer, pre-clean and immunoprecipitation (incubation with antibodies and beads) were performed as described above in the ChIP section, except Pierce Protein A/G Magnetic Beads (Thermo Scientific; 88802) were used instead of Protein G Plus/Protein A beads. Prior to adding antibodies, the pooled chromatin derived from each genetic/tissue compartment was divided into two for E2F3- and MYC-ChIP, respectively. After generating the chromatin-antibody-bead complex, the library construction steps including on-bead enzymatic reactions (end polishing, P7 exo-adapter ligation, nick repair, λ-exonuclease digestion, RecJf exonuclease digestion), elution and reverse-crosslinking, primer extension and P5 exo-adaptor ligation were performed as described11. The resulting DNA was enriched by 13 cycles of PCR using NEBNext High-Fidelity PCR Master Mix (NEB; M0541S). The DNA concentration and DNA size distribution of libraries were profiled using Agilent High Sensitivity DNA Kit. Cluster generation on Illumina cBot and single-end high throughput sequencing on Illumina HiSeq 2500 platform were performed at the Ohio State University Shared Resources (http://cancer.osu.edu/research-and-education/shared-resources/genomics/services).
Microarray data analysis
For microarray analysis, functions from R package Affy12 were utilized. The ‘justRMA’ function was used to read the raw CEL files, perform background correction and normalization. The ‘exprs’ function was used to obtain the log2 transformed expression value. All parameters were set as default.
ChIP-exo-seq data processing
After base calling, adapter trimming, and barcode demultiplexing using the sequencer manufacturer’s software, raw sequences with quality scores (Sanger/Illumin1.9) were stored in FASTQ format. The name of each sample includes the mouse genotype, tissue type and the specific antibody used. Each sample was sequenced in two lanes of an Illumina flow cell and the resulting data was combined into one FASTQ file before mapping onto the genome.
We used Bowtie2 (ref. 13) to align short reads onto the mouse genome (mm9) with -N 1 to allow only one mismatch and output alignments into SAM formatted files using -S option. Two algorithms were then used with the default significance cutoffs (GEM7: FDR<0.01; MACS2 (ref. 5): FDR<0.05) to detect peaks enriched on the genome. Any binding events identified by GEM that were not identified by MACS2 were filtered out by using ‘window’ command from bedtools14 with -w 1000. We used GEM to predict binding positions and MACS2 to filter binding events because GEM has a higher accuracy of predicting binding locations and binding motifs. To predict genes regulated by these binding events, ‘refgene_getnearestgene’ command from CisGenome15 was used to find the nearest gene for each binding event. We set -dt 1, -up 50,000,000 and -down 30,000,000 so that binding events located 50 M bp upstream or 30 M bp downstream of the nearest TSS were discarded. The gene annotation file we used is mm9_refFlat_sorted.txt, which was downloaded from CisGenome website. Next, we used HOMER16 script findMotifsGenome.pl with -size 100 to do de novo searches of enriched sequence motifs within 100 bp regions of the binding events. Finally, HOMER script annotatePeaks.pl was used to calculate read coverage over the 500 bp regions of binding events and HOMER script Homer_plot.R was used to visualize the read coverage.
Code availability
The version of each software used in this paper are noted below.
bedtools v2.17.0
samtools 0.1.19-44428 cd
FastQC v0.11.2
bowtie2 version 2.2.1
macs2 2.0.10.20131216 (tag:beta)
GEM (version 2.4.1)
homer v4.6
IGV Version 2.3.32
All the scripts used to run tools mentioned before or process intermediate files are uploaded onto figshare (https://dx.doi.org/10.6084/m9.figshare.2059239)
Data Records
ChIP-exo-seq data from 8 samples (4 from crypts and 4 from villi) were deposited in Gene Expression Omnibus17 (NCBI) under accession number GSE56008 (Data Citation 1). For each sample in GSE56008, a processed data file listing all the predicted binding sites is provided. It is a tab-separated text file in BED format with the following columns: Chromosome Name, Start Coordinate, End Coordinate, Binding Site ID, and Binding Site Score (calculated from GEM). Links to the corresponding NCBI Sequence Read Archive (SRA) accession, which deposits the raw sequencing data (Data Citation 2), are also provided.
Microarray Dataset 1 in Fig. 2 were deposited in GEO under the accession number GSE56006 (Data Citation 3). Microarray Dataset 2 in Fig. 2 were deposited in GEO under accession number GSE56007 (Data Citation 4). Raw signal files in.CEL format are also provided for each sample. Users could freely download them and reanalyze them with R package Affy.
Technical Validation
ChIP-exo-seq
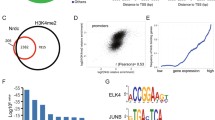
To survey the quality of our ChIP-exo-seq data the average quality score for each nucleotide position was determined (Sanger/Illumina1.9 format). Figure 3a shows an example of the quality score for each position (bp) in tag reads for the E2F3-specific ChIP-exo-seq data set derived from Rb deleted villi. In all ChIP-exo-seq experiments the quality score was determined to be above 30 (Fig. 3a). Over 90% of the sequenced reads were mapped successfully to the mouse genome (mm9), where over 60% of reads mapped to unique genome positions (Table 2). Thus, based on nucleotide position quality score and the percentage of reads mapped to the genome, we conclude that the sequencing quality of our ChIP-exo-seq data is very good.
(a) Bar plot for bases quality score from a representative sample ‘RbKO villi anti-E2F3’. Tag (5 prime end of read) distribution surrounding predicted binding events on average from representative samples (b) ‘RbKO villi anti-E2F3’ and (c) ‘RbKO villi anti-MYC’. Tags from each DNA strand are shown both separately (blue: +; red:−) and combined together (yellow) on the same plot.
Reads aligned to the positive and negative strands formed independent single peak summits for both E2F3 and MYC ChIP experiments. Sequences between the two summits, which were 24 bp–28 bp in length, represent the E2F3 and MYC binding sites (Fig. 3b and c). The number of reads between the two summits were >19-fold higher than in other genomic regions (Table 3). Thus, ChIP-exo-seq derived reads sharply define the protein binding sites of E2F3 and MYC on chromatin.
To validate the predicted transcription binding sites, forty randomly selected E2F3- and MYC-specific peaks were assessed by ChIP-PCR assays in E2f3-deficient and Myc-deficient tissues, respectively. All selected E2F3- and MYC-specific peaks were highly enriched in wild type tissues when compared to E2f3- and Myc-deficient tissues (see NCB paper9 Fig. 4b). In addition, control IgG ChIP-PCR assays showed low background binding to these selected E2F3- and MYC binding sites. Together, these control experiments highlight the highly specific nature of E2F3- and MYC chromatin profiling by ChIP-exo-seq.
We then identified transcription factor binding motifs imbedded within 100 bp region of the identified binding sites. As expected, canonical E2F binding motifs were significantly enriched in crypts and villi of both wild type and Rb deficient tissues. Interestingly, canonical MYC binding motifs were only enriched in Rb deficient crypts, highlighting the impact of Rb loss on MYC global chromatin binding. Detailed information of motif sequences and distribution for each experiment is listed in Supplementary Fig. 6b of the reference manuscript9.
Expression profiling
Microarray gene expression data sets were first background corrected and normalized, and then the log2 transformed expression values were used to perform Multidimensional Scaling (MDS) analysis. As shown in Fig. 2, replicate samples within the same genetic cohort grouped together. Finally, gene expression changes between genetic cohorts were validated by real-time PCR expression assays (Figs 3b, 3c, 6d and Supplementary Fig. 3 of Liu et al.,9). Thus, we conclude that the overall reproducibility of microarray gene expression data sets is very good.
Usage Notes
Scripts for running Bowtie2, GEM, MACS2, HOMER and scripts for processing other intermediate files have been uploaded to figshare (https://dx.doi.org/10.6084/m9.figshare.2059239). These scripts were run on a linux system. To deploy the pipeline, please download and decompress the script archives, install required software and configure them in the ‘software.conf’ file. Then copy your raw FASTQ files into the ‘rawdata’ folder and configure them in the ‘samples.conf’ file. Finally, the ‘bash run_ChIP-exo_pipeline.sh’ command is used to execute the analysis pipeline. The predicted binding regions, corresponding target genes, and enriched sequence motifs of each ChIP-exo-seq experiment will be automatically generated. Intermediate files, such as alignments in BAM format and read coverage along the genome, will also be generated and could be visualized using IGV18. The parameters can be adjusted by revising the corresponding scripts. This package may be modified to accommodate other species.
Additional Information
How to cite this article: Tang, X. et al. Transcriptome regulation and chromatin occupancy by E2F3 and MYC in mice. Sci. Data 3:160008 doi: 10.1038/sdata.2016.8 (2016).
References
References
Chen, H.-Z., Tsai, S.-Y. & Leone, G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nature Reviews Cancer 9, 785–797 (2009).
Amati, B., Alevizopoulos, K. & Vlach, J. Myc and the cell cycle. Front Biosci. 3, d250–d268 (1998).
Meyer, N. & Penn, L. Z. Reflecting on 25 years with MYC. Nature Reviews Cancer 8, 976–990 (2008).
Crosnier, C., Stamataki, D. & Lewis, J. Organizing cell renewal in the intestine: stem cells, signals and combinatorial control. Nature Reviews Genetics 7, 349–359 (2006).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome biology 9, R137 (2008).
Schmidt, D. et al. ChIP-seq: Using high-throughput sequencing to discover protein–DNA interactions. Methods 48, 240–248 (2009).
Guo, Y., Mahony, S. & Gifford, D. K. High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Comput. Biol. 8, e1002638–e1002638 (2012).
Rhee, H. S. & Pugh, B. F. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 147, 1408–1419 (2011).
Liu, H. et al. Redeployment of Myc and E2f1-3 drives Rb-deficient cell cycles. Nat. Cell Biol. 17, 1036–1048 (2015).
Ireland, H. et al. Inducible Cre-mediated control of gene expression in the murine gastrointestinal tract: effect of loss of β-catenin. Gastroenterology 126, 1236–1246 (2004).
Serandour, A. A., Brown, G. D., Cohen, J. D. & Carroll, J. S. Development of an Illumina-based ChIP-exonuclease method provides insight into FoxA1-DNA binding properties. Genome Biol. 14, R147 (2013).
Gautier, L., Cope, L., Bolstad, B. M. & Irizarry, R. A. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315 (2004).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nature methods 9, 357–359 (2012).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Ji, H., Jiang, H., Ma, W. & Wong, W. H. Using CisGenome to Analyze ChIP‐chip and ChIP‐seq Data. Current Protocols in Bioinformatics 2.13.11–12.13, 45 (2011).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell 38, 576–589 (2010).
Edgar, R., Domrachev, M. & Lash, A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research 30, 207–210 (2002).
Thorvaldsdóttir, H., Robinson, J. T. & Mesirov, J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in bioinformatics 14, 178–192 (2012).
Data Citations
Acknowledgements
This dataset was generated for the referenced manuscript by Liu et al. The authors would like to thank Sooin Bae for advice on sequencing library construction and Nick Selner for assistance with editing the manuscript. This work was supported by the Genomics Shared Resource (GSR) at Ohio State University and Ohio Supercomputer Center. This work was funded by NIH grants to G.L. (R01CA121275 and R01HD047470) and NIH grant to J.M.P. (R01CA098956).
Author information
Authors and Affiliations
Contributions
H.L. and G.L. designed the experiments. H.L. performed the wet-lab experiments. X.T., A.S., T.P., P.C., performed the bioinformatics and statistical analysis of gene expression and ChIP-exo-seq data sets with supervision from K.H., R.M. and G.L. Z.C. and Q.W. provided guidance on the ChIP-exo-seq experiments. J.M.P. and G.L. supervised the overall study. X.T., H.L. and G.L. wrote the manuscript with inputs from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interest.
ISA-Tab metadata
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0 Metadata associated with this Data Descriptor is available at http://www.nature.com/sdata/ and is released under the CC0 waiver to maximize reuse.
About this article
Cite this article
Tang, X., Liu, H., Srivastava, A. et al. Transcriptome regulation and chromatin occupancy by E2F3 and MYC in mice. Sci Data 3, 160008 (2016). https://doi.org/10.1038/sdata.2016.8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/sdata.2016.8
- Springer Nature Limited
This article is cited by
-
Directed differentiation of human pluripotent stem cells into diverse organ-specific mesenchyme of the digestive and respiratory systems
Nature Protocols (2022)
-
Single cell transcriptomics identifies a signaling network coordinating endoderm and mesoderm diversification during foregut organogenesis
Nature Communications (2020)