Abstract
Fungi constitute the Earth’s second most diverse kingdom, however only a small percentage of these have been thoroughly examined and categorized for their secondary metabolites, which still limits our understanding of the ecological chemical and pharmacological potential of fungi. In this study, we explored members of the co-evolved termite-associated fungal genus Xylaria and identified a family of highly oxygenated polyketide-terpene hybrid natural products using an MS/MS molecular networking-based dereplication approach. Overall, we isolated six no yet reported xylasporin derivatives, of which xylasporin A (1) features a rare cyclic-carbonate moiety. Extensive comparative spectrometric (HRMS2) and spectroscopic (1D and 2D NMR) studies allowed to determine the relative configuration across the xylasporin family, which was supported by chemical shift calculations of more than 50 stereoisomers and DP4+ probability analyses. The absolute configuration of xylasporin A (1) was also proposed based on TDDFT-ECD calculations. Additionally, we were able to revise the relative and absolute configurations of co-secreted xylacremolide B produced by single x-ray crystallography. Comparative genomic and transcriptomic analysis allowed us to deduce the putative biosynthetic assembly line of xylasporins in the producer strain X802, and could guide future engineering efforts of the biosynthetic pathway.
Similar content being viewed by others
Introduction
The fungal kingdom ranks as the second most diverse kingdom on Earth, estimated to comprise 3–4 million species1. Within this kingdom, Sordariomycetes stands out as a diverse and significant class, including fungal families such as Hypoxylaceae and Xylariaceae1,2 which have recently gained considerable attention due to their enormous biosynthetic repertoire to produce secondary metabolites with diverse biological activities3,4, While the Xylariaceae family includes predominately free-living saprophytic members that are often noticeable due to the development of stromatal tissue on decaying wood, it also includes a distinctive Xylaria subgenus, previously termed Pseudoxylaria, whose member have been isolated from fungal garden material of fungus-farming termites, in which termites cultivate the fungal mutualist Termitomyces as a crop5,6,7, Members of the co-evolved Xylaria subgenus exclusively appear in the fungus comb of termite colonies that are weakened or abandoned, rapidly dominating any remaining termite food source (Fig. 1)8,9,
In our recent investigation, we delved into the genomic and biochemical foundations of the distinctive co-evolved antagonistic behavior and substrate specialization of termite-associated strains10, and posited that the secreted antimicrobials could play a role in outcompeting the fungal mutualist once vegetative mycelium emerges11,12,13,14,15. Transcriptomic and metabolomic studies indeed showed that metabolite production is triggered and potentially diversified when termite-associated strains were cultivated on comb-mimicking growth conditions and in fungus-fungus co-cultivations against the fungal cultivar10,16. Furthermore, the chemical nature of secreted metabolites differed across the fungal isolates, which was also reflected in the isolate-specific genetically encoded biosynthetic repertoire. Especially, a biosynthetic gene cluster (BGC) region assigned to the cytosporin/xylasporin family caught our attention as the core BGC was encoded in the genome of four out of seven different termite-associated isolates, but variations in the abundance and arrangement of accessory genes was detectable. While genes coding for a short chain oxidoreductase (pxB; SDR) and a cupin protein (pxF) were lacking in the genomes of isolate X187, Mn153, and Mn132, the additional SnoaL-like polyketide cyclase (pxP) was present in X187, Mn153 and Mn132, and only lacking in isolate X80211. Therefore, we postulated the existence of a previously undiscovered level of structural diversity within the xylasporin/cytosporin family produced by strain X802, in contrast to those isolated from X18710. To further solidify the hypothesis and clarify the chemical nature of the cytosporin/xylasporins produced by the other termite-associated fungal isolates, we re-investigated the secondary metabolome of the model strain Xylaria sp. X8028,11 using eco-mimetic cultivation conditions and a comparative HRMS2-based metabolomic approach.
Here, we report the isolation and characterization of six yet unreported polyketide-terpene hybrid metabolites named xylasporins A-F (1–6) from Xylaria sp. X802 and the full structure determination of previously identified xylasporin I (8) from Xylaria sp. X187. Due to the structural complexity of this compound class, we pursued a comprehensive comparative NMR study coupled with chemical-computational calculation of more than 50 possible stereoisomers to enable the deduction of its relative and absolute stereochemistry. Furthermore, we were able to obtain single x-ray data of the commonly co-secreted polyketide-NRPS hybrid metabolite, xylacremolide B (9), enabling a revision of its stereochemistry. 14 Our comparative genomic studies also allowed deducing the putative polyketide-terpene-based biosynthetic origin.
Results and discussion
Cultivation and comparative analysis
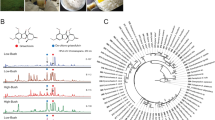
To study if ecomimetic conditions could trigger the production of the cytosporin/xylasporin-specific features, Xylaria sp. X8028,11 was co-cultivated with the fungal mutualist Termitomyces sp. T153 (7–14 days, 30 °C) (Fig. 2a)10. Culture extracts were analyzed for (induced) metabolite production by comparing the metabolome of the interaction zone (ZOI) to metabolites secreted from an axenic agar culture grown on potato-dextrose agar (PDA) and previous experimental set-ups10 using liquid chromatography (LC) coupled with tandem electron-spray ionization (ESI)-HRMS/MS analyses. Obtained MS2-data was dereplicated using the Global Natural Product Social Molecular Networking Web platform (GNPS) (Supplementary Fig. S1)17. The comparative molecular network revealed several molecular ion clusters containing more than ten different nodes, including a cluster putatively assigned to diketopiperazines, a cluster assigned to previously identified pseudoxylallemycins (m/z 549.322, 617.456, 619.262, and 685.394)11, and a cluster assigned to cytochalasin derivatives (m/z 524.263) (Supplementary Fig. S2)11. However, the MS2-cluster that was tentatively assigned to the cytosporin/xylasporin family from Xylaria sp. X802 included several spectral nodes with yet unknown molecular ion features, and which differed from those observed in the GNPS-subcluster of isolate X187 (Fig. 2b)10.
X802 grown on different media and co-culture conditions. a Co-cultures of Xylaria sp. X802 versus Termitomyces sp. T153 on PDA/soil medium; (b) Expanded GNPS-based molecular network cluster of xylasporin-based molecular ion features. Features that were only present in the zone of inhibition (ZOI) are of pink color and those from axenic growth of X802 are colored in blue (1, m/z 337.164 [M-H2O + H]+, 2/3, m/z 293.174 [M-H2O + H]+, and 4/5/6 m/z 311.184 [M-H2O + H]+). c Culture of Xylaria sp. X802 on rice-sawdust medium, and (d) glass beads with liquid PDB. e Selected ion chromatogram of 1 (m/z = 337.1636 [M-H2O + H]+), 2/3 (m/z = 293.1740 [M-H2O + H]+), 4/5/6 (m/z = 311.1847 [M-H2O + H]+) and 7 (m/z = 193.1223 [M + H]+) with extracts derived from (ii) agar plate cultivation, (iv) glass beads cultivation with liquid PDB and (vi) rice-sawdust medium of three weeks old Xylaria sp. X802. The corresponding negative controls without X802 are shown below each lane (i, iii and v), respectively.
We then investigated the abundances of cytosporin/xylasporin-specific features within culture extracts obtained from cultures grown on rice-sawdust medium or on glass beads (Fig. 2c, d)10. While the cytosporin/xylasporin-specific molecular ion features were detectable from Xylaria sp. X802 within all different cultivation conditions (Fig. 2e), some features were only detectable in samples derived from the interaction zone of a co-culture experiment (Fig. 2b). In addition, the intensity of each molecular ion feature was depended on the substrate provided for growth and was indicative for subtle transcriptional regulations of tailoring enzymatic activity as indicated by prior studies10.
Structural elucidation of xylasporin derivatives
We then pursued the isolation of the unknown xylasporin-specific chemical features from isolate X8028,10,11. For this, Xylaria sp. X802 was grown for 14 days on PDA plates and mycelium-covered plates were then extracted with methanol to yield an enriched crude extract. Compounds were purified by MS- and UV-guided reverse-phase chromatography. Overall, we were able to isolate seven metabolites. Comparative NMR analysis of compounds 1–6 uncovered a similar 1D and 2D NMR pattern, which indicated towards a shared bicyclic, and oxidized core skeleton (Tables S1 and S2). In addition, a known biosynthetic precursor, 1-hydroxy-2-hydroxymethyl-3-pent-1-enylbenzene (7) was characterized according to previous data.
The molecular formula of compound 1, C18H26O7, was deduced from HRESI( + )MS data [m/z 709.3430 [2 M + H]+, calcd. for C36H53O14, 709.3435]. The 1H NMR data of 1 showed the presence of signals for three methyl groups, three methylene units, one oxygenated methylene, four oxygenated methines, and two olefinic protons (Tables S1 and S2). HSQC and HMBC spectra revealed a total of 18 carbon signals attributable to three methyls, four methylenes, six methines, four quaternary carbons, and one carbonyl-like carbon (Table S1). Interpretation of 2D NMR spectra allowed the determination of planar structure of 1, with notable HMBC correlations between H-7/C-14, and H-6/C-19 and H-7/C-19 showing the presence of a cyclic carbonate group, as well as COSY correlations of an aliphatic spin system (Fig. 3a). Overall compound 1 showed similarities to cytosporin E featuring a pentenyl chain at C-8 compared to a heptenyl chain in cytosporin E (Fig. 3c)18. The relative configuration of 1 was determined by vicinal 1H-1H coupling constants (3JHH) and ROESY correlations of H-3/H3-12, H-10/H3-11, H3-11/H-4α, and H-6/H-4β indicating the equatorial orientation of the hydroxyl group at C-3 and the axial orientation of H-6, H-7, and H-10 (Fig. 3a). As the hexahydrobenzopyrane moiety of compound 1 showed NMR chemical shifts very close to those of cystoporin E, we deduced that these compounds share the same absolute configuration: 3 S, 5 R, 6 R, 7 R, and 10 S (Supplementary Table S3, Supplementary Figs. S3–S8).To solidify our structural assumptions, we also calculated the 1H and 13C shifts using optimized settings for Gauge-including atomic orbital (GIAO) NMR chemical shifts calculations, which was followed by DP4+ probability analysis19 of overall 50 stereoisomers of the three structure types that encompass compound 1–6 (Supplementary Tables S11–S22). This effort allowed us to confirm the cis-orientation of the angular substituents at C-5 and C-10 of compound 1 (Supplementary Tables S4 and S5, Supplementary Fig. S9).
a Planar structure of xylasporin A (1) and key 2D NMR correlations for compound 1; (b) Experimental ECD spectrum for compound 1 and Boltzmann-averaged TD-DFT calculated ECD spectra for 1a (3S,5R,6R,7R,10S), 1b (3R,5S,6S,7S,10R); (c) Proposed absolute structure of xylasporin A (1) and reported cytosporin E. d Structures of known carbamate-containing fungal metabolites phomoxins and aspergillusols sharing similar structural and biosynthetic features.
Additionally, we compared the experimental ECD spectrum of 1, which exhibits two positive Cotton effects at 228 nm and 265 nm, with the calculated ECD spectra for the most stable conformers of the two possible enantiomers of 1 [1a (3S,5R,6R,7R,10S) and 1b (3R,5S,6R,7S,10R)], which in summary allowed us to deduce the absolute configuration for compound 1 (3S,5R,6R,7R,10S) (Fig. 3b). Thus, we named compound 1 xylasporin A due to its structural resemblance to the cytosporin family20,21,22,23,24,25,26,27,28,29, Here, it was intriguing to note that cyclic-carbonate patterns are rarely found in natural products, and have so far predominately been reported from the marine-derived fungus Peroneutypa scoparia (previously termed Eutypella scoparia)20, Aspergillus sp. PSU-RSPG185 (aspergillusols)23, and Phoma sp. (phomoxins) (Fig. 3c)30.
The molecular formula of xylasporins 2 and 3 was determined as C17H26O5 based on the HRESI(+)MS analyses, which showed pseudo-molecular ions peaks at m/z 293.1742 [M-H2O + H]+ (calcd. for C17H25O4, 293.1753) for 2 and m/z 621.3627 [2 M + H]+ (calcd. for C34H53O10, 621.3639) for 3, indicating that these compounds are likely structural isomers. The 1D and 2D NMR spectra of 2 and 3 exhibited some similarities with those signals assigned to the core structure of 1. However, signals for the carbamate group were lacking and chemical shift patterns at position C-5, C-6, and C-7 were suggestive for the presence of an epoxide group at C-5 and C-6 (Fig. 4a, b, Tables S1 and S2). Comparison with literature data suggested that compounds 2 and 3 had a similar planar structure to cytosporins D and M, but carried a pentenyl chain at C-8 in contrast to a heptenyl chain in cytosporins D and M (Fig. 4c)18,27. Differences in chemical shifts and 3JHH coupling constants for H2-4, as well as key ROESY correlations supported the notion that compounds 2 and 3 share the same relative configurations 3 S*, 5 R* and 6 S* with an inversion of relative configurations for C-7 and C-10 (2: 7 R*, 10 S*; 3: 7 S*, 10 R*) (Fig. 4a, b). Comparison of 1H and 13C NMR chemical shifts of the hexahydrobenzopyrane moieties of compound 2 and cytosporin D led us to deduce the putative absolute configuration of 2 as 3 S, 5 R, 6 S, 7 R, and 10 S [(\(\bar{{{{{{\rm{X}}}}}}}\)(|ΔδH(2-cystoporin D) | ) = 0.04 ppm, s = ΔδH(2-cystoporin D) = 0.05 ppm; \(\bar{{{{{{\rm{X}}}}}}}\)(|ΔδC(2-cystoporin D) | ) = 0.24 ppm, s = ΔδC(2-cystoporin D) = 0.15 ppm)] (Fig. 4a, Supplementary Tables S6–S9, Supplementary Figs. S10–S22)18. These deductions were solidified by GIAO NMR chemical shifts calculation followed by DP4+ probability, which additionally allowed us to propose the relative configuration for xylasporin C (3) as 3S*,5R*,6S*,7S*, and 10R* (Supplementary Table S10–S13, Supplementary Figs. S22 and S23).
a Proposed structure of xylasporin B (2), structure with |ΔδC(2-cystoporin D)| values in ppm and 3D depiction showing ROESY correlations; (b) Proposed structure of xylasporin C (3), structure with |ΔδC(3-cystoporin D)| values and 3D depiction showing ROESY correlations; (c) Structures of reported natural products with similar core scaffolds used for comparison.
The molecular formulas of xylasporins D (4), E (5), and F (6) were deduced from the sodium-adducted ion peak at m/z 351.1784 [M+Na]+ (calcd. for C17H28O6Na, 351.1784) from HRESI(+)MS data of 4, and from the pseudo-molecular ions peaks at m/z 657.3850 [2 M + H]+ and m/z 657.3855 [2 M + H]+ (calcd. for C34H57O12, 657.3850) from HRESI( + )MS data of 5 and 6.
Interpretation of the 1H and 13C NMR spectra of these compounds revealed again the hexahydrobenzopyrane scaffold with chemical shift differences at C-5, C-6, and C-7 compared to 1, which suggested the presence of hydroxyl groups at C-5, C-6, and C-7. COSY and key HMBC correlation analyses allowed deducing the planar structures of 4, 5, and 6, resembling that of cytosporin L, which includes a pentenyl chain at C-8 compared to a heptenyl chain in cytosporin L (Fig. 5,)3. Possible relative configurations of C-3, C-5, C-6, C-7, and C-10 of 4 were deduced from key ROESY correlations (Fig. 5b, Supplementary Figs. S25–S43) and GIAO NMR chemical shifts calculations of a reduced set of possible stereoisomers followed by DP4+ probability all together supported the proposed configuration of 3 S*, 5 S*, 6 R*, 7 R*, and 10 R* for compound 4 (Supplementary Tables S14 and S15, Supplementary Fig. S30).
We then compared the 1H and 13C chemical shifts for compound 5 and 6 and found that the chemical shift values of C-6 and C-7, and the vicinal coupling constant between the protons H-6 and H-7 (4: 6.5 Hz vs. 5 and 6: 4.5 Hz) in relation to those values obtained for compound 4 were suggestive for an S-stereochemistry at position C-6 and C-7. This deduction was also supported by key ROESY correlations (H-6/H-10 for 5 and H-6/H-4β for 6). Stereochemistry at position C-10 was deduced for both compounds from signatory 13C chemical shift patterns and ROESY correlations (5: H-10/H-3, H-3/H3-12, and H3-12/H-4α; 6: H-10/H-4α and H-4α/H3-11) (Fig. 5b). The stereochemical configuration of C-5 in compound 5 remained an open question as the experimental and calculated NMR data resulting of DP4+ probability of 78% (Supplementary Tables S16 and S17, Supplementary Fig. S37). While still ambiguous, we propose that ROESY correlations between H-10 and H-3, and H-10 and H-6 are suggestive for a 5 S configuration for compound 5. In contrast, the stereochemistry at C-5 for compound 6, was determined based on the key H-10/H-4α ROESY correlation and unambiguous DP4+ analysis of 100% (Fig. 5B, Supplementary Tables S18 and S19, Supplementary Fig. S44). Overall, the combination of NMR interpretation and calculation allowed us to proposed the relative configuration for 5 as 3 S*,5 S*,6 S*,7 S*,10 R* and for 6 as 3 S*,5 R*,6 S*,7 S*,10 S*.
Finally, we also compared the 1H and 13C shifts using optimized settings for GIAO-NMR chemical shifts calculations of several stereoisomers across each structure type (Tables S4–S5, S10–S13, S14–S19). Overall, we were able to deduce patterns (“rule of thumbs”) that correlated to the stereochemistry of C-3 and C-10 and the 13C chemical shifts of C-11 and C-12. In case the same stereochemistry (S, S or R, R) was observed for C-3 and C-10, the 13C chemical shifts of C-11 and C-12 differed by more than 10 ppm. In case the stereochemistry of C-3 and C-10 were inverted (R, S or S, R), the 13C chemical shift values for C-11 and C-12 differed less than 3 ppm.
Relative structure elucidation of xylasporin I and structure revision of xylacremolide B from isolate X187
Prior comparative genomic and metabolomic analyses proposed that Xylaria strains carrying fog-like gene clusters should produce xylasporin-like metabolites, which led to first isolation efforts and deduction of the planar structure of two congeners xylasporin H and I from strain X18710. However, due to the light- and acid-sensitive chemical features of the conjugated polyene system, only limited analytical data was obtained leaving the relative stereochemistry of these congeners unassigned. Building up on the acquired analytical and calculated NMR data of the xylasporin-compound class within this study, we herein addressed again the intrinsic challenge of assigning the relative stereochemistry to isolates of this compound family. For re-isolation, agar plates covered with fungal mycelium of Xylaria sp. X187 were used, which yielded after ethyl acetated-based extraction, xylasporin I (8) in 1.2 mg alongside with the co-secreted natural products xylacremolides A and B (9) (Fig. 6)10,14. Despite the instability of xylasporin I (8), we were able to obtain this time sufficient quality of 2D-NMR data for structural and computational analysis (Supplementary Table S20, Supplementary Figs. S48–54). 2D NMR analysis confirmed that xylasporin I (8) shares the core structure and chain length with xylasporins B (2) and C (3) produced by Xylaria sp. X802; however, it was distinguished by a pentdienyl chain instead of the heptenyl chain present in cytosporins D and M18,27. Comparative NMR studies and GIAO NMR chemical shifts calculations using optimized settings (Supplementary Tables S21–S22, Supplementary Fig. S55) allowed us to propose the relative stereochemistry for xylasporin I (8) as 7 R*, 8 S*, 9 R*, 11 S*, 15 R* (Fig. 6a).
a Chemical structure of xylasporin I (8) and its 3D depiction showing ROESY correlations. b Revised chemical structure of xylacremolide B (9), crystal structure of xylacremolide B (9) shown as ORTEP plot of with displacement ellipsoids of non-hydrogen atoms drawn at 50% probability level and chemical structure of xylacremolide A14.
In light of our efforts to obtain single crystals for any of the secondary metabolites isolated from strains X802 and X187 to validate the deductions from chemical shift calculations, only purified xylacremolide B (9) crystallized from a mixture of methanol and water (Fig. 6b, Supplementary Table S24, Supplementary Fig. S61).
In combination with our previous reports, which included the determination of the relative stereochemistry (d-Phe and l-Pro) of xylacremolides A and B (9) using Marfey’s method and a single crystal X-ray analysis of xylacremolide A, we were able to serendipitously revise the chemical structure of xylacremolide B (9) (Fig. 6b)14. Single X-ray analysis of 9 uncovered two important features: firstly, the observed orientation of the 2-CH3 towards the aromatic ring of Phe indeed leads to a direct exposure to the ring current, likely causing the significant upfield-shift of the chemical shift assigned to 2-CH314. Secondly, while previous NOESY interpretation suggested a 2 S,3 R,5R-configuration of the PKS-derived hydroxylated alkyl chain, single X-ray analysis uncovered a 2 R,3 R,5R-configuration, and thus let to a revision of the relative and absolute configuration of xylacremolide B (9).
Biosynthetic considerations
Cytosporins and xylasporins share the same planar hexahydrobenzopyran backbone, with xylasporins carrying a pentenyl chain at C-8 compared to a heptenyl chain in cytosporins. As the planar hexahydrobenzopyran backbone suggested a mixed polyketide and terpene-type biosynthetic origin18,19,20,21,22,23,24,25,26,27,28,29,30,31, we revisited our previous comparative genome analysis of Xylaria genomes10, which reported four strains (X802, Mn132, Mn153, and X187) to encode a cluster region bearing resemblance to the fog BGC (Aspergillus ruber)4 responsible for the biosynthesis of glaucin-type of scaffolds. The locus within the genome of Xylaria sp. X802 was assigned the px gene cluster (BGC) region (Fig. 7, Supplementary Table S25), which encompasses a highly-reducing poly ketide synthase PKS (HR-PKS) pxA (KS-AT-DH-ER-KR-ACP), an ABBA type aromatic prenyltransferase pxH, three short-chain dehydrogenases/reductases (SDR) pxB/pxD/pxJ, two cupins pxC/pxF, a cytochrome p450 monooxygenases (CYP) pxE, two FAD-dependent oxidoreductases pxG/pxI, two fungal transcription factors pxK/pxM, a major-facilitator superfamily (MFS) transporter pxL, a phosphopantetheinyl transferase pxN and a thioesterase domain containing enzyme pxO32.
a Schematic depiction of the px BGC in Xylaria sp. X802 and comparison to homologs BGCs from the biosynthesis of flavoglaucin (Aspergillus ruber CBS 135680), trichoxide (Trichoderma virens sp. Gv29-8) and sordarial (Neurospora crassa sp. OR74A). Shown are genes coding for highly-reducing polyketide synthases (blue; HR-PKS), short-chain dehydrogenases/reductases (red; SDR), cupins (silver), cytochrome p450 monooxygenases (green; CYP), flavin dependent oxidoreductases (yellow; OR), aromatic prenyltransferases (purple; PT), transcription factors (black) and major facilitator superfamily transporters (dark gray; MFS). Non-related genes are shown in light gray. b Putative biosynthetic assembly line yielding isolated biosynthetic precursor 7, which is likely biotransformed through a series of oxidative transformation steps to yield epoxidized xylasporins of type 2/3, tetradiols 4-6 and in a yet unidentified step carbonate 1.
A detailed in silico of the px BCG uncovered that seven of the encoded proteins (PxA-E and PxG-I) showed high similarity (42–86% similar amino acid sequence; Supplementary Table S20) to the characterized FogA-F from Aspergillus ruber sp. CBS 135680, which is responsible for the biosynthesis of the prenylated salicylic acid derivative flavoglaucin31. Homologs gene sequences were also previously identified in other ascomycetes like Trichoderma virens sp. Gv29-8 (61–80%)33 and Neurospora crassa sp. OR74A (47–76%)34, encoding for the biosynthesis of the prenylated PKS-based salicylic acid derivatives sordarial and trichoxide, respectively. Based on these homologies, an iterative incorporation of five malonyl-CoA onto an acetyl-CoA starter unit by the HRPKS PxA was proposed to yield ACP-bound intermediate I. Here, it is important to point out that xylasporins A-F (1–6) are characterized by a pentenyl chain at C-8 compared to a heptenyl chain in cytosporins, which suggested yet unknown structural and functional difference in the iterative mode of the HRPKS PxA compared to the fogA-PKS reported in Aspergillus.
The strain-specific modifications of the polyketide chain should be realized by the KR, DH, and ER domains of the HR-PKS PxA as well as the short-chain dehydrogenases/reductase PxB and cupin PxC, similar to mechanisms that have been shown for flavoglaucin biosynthesis in A. ruber31. Release of the polyketide II is proposed to be catalyzed by SDR PxD or the putative trans-acting TE domain containing protein PxO leading to the production of the detectable precursor metabolite 7. Hydroxylation of compound 7 at position C-3 by PxE and prenylation at C-5 by PxH could lead to the synthesis of a flavoglaucin congener (IV). The double bond at the prenyl group should then be epoxidized by either the putative cytochrome P450 monooxygenases PxF or PxG. The chromene core structure of VI is likely formed by spontaneous ring closure leading to two distinct stereoisomers (m/z = 275.164 [M-H2O + H]+). Subsequent oxidation of the alcohol at C-7 by either PxI or PxJ might form an unsaturated enone VIII, while a second epoxidation by PxF or PxG could catalyze the formation of 2 and 3. Enzymatic or spontaneous hydrolysis of the oxyrane ring and reduction of the carbonyl functionality should provide the three isomers 4–6, but the enzymatic basis for the regioselective introduction of the cyclic carbonate moiety still remains enigmatic.
Antibacterial activity studies
Cytosporins A-C, isolated from an endophytic strain of the taxonomically related fungus Cytospora5, were reported to act as angiotensin II binding inhibitor, while cytosporin L was shown to have activity against the bacteria Micrococcus lysodeikticus and Enterobacter aerogenes with MIC values of 3.12 µM18. However, cytosporin D and E were reported to have neither antibacterial nor antifungal activity against Staphylococcus aureus, Escherichia coli, and Candida albicans23,27.
Indeed, when we evaluated the antibacterial activity of stable xylasporins derivatives A-F (1–6) against a panel of bacterial test strains, neither of the derivatives exhibited antibacterial activity. As xylasporins A-F (1–6) were detectable in the ZOI of the fungus-fungus co-culture (Fig. 2), their antifungal activity against A. nidulans RMS011 as test strain was evaluated. However, only very mild antifungal activity in form of spore germination inhibition was observed for xylasporins B (2) and C (3) to a similar extend as the co-isolated 19,20-epoxycytochalasin Q and pseudoxylallemycin A (Supplementary Fig. S62)11.
Conclusion
Members of the termite-associated subgenus of Xylaria sp. are substrate-specialized and emerge from abandoned fungus comb material of fungus-growing termites. Building on the discovery that termite-associated strains inhibit the growth of the fungal crop Termitomyces assisted by its biosynthetic capabilities to produce secondary metabolites, we substantiated the hypothesis by conducting experiments under eco-mimetic growth conditions and employing a comparative HRMS2-based metabolomic approach. Dereplication of metabolic features, whose abundances were dependent on the provide growth conditions, led to the discovery of six yet unreported secondary metabolites of PKS-terpenoid origin from strain X802, which we named xylasporins A-F (1–6) due to their hexahydrobenzopyran backbone and structural resemblance to cytosporins, and the reisolation of the instable xylasporin I (8) from strain X187. GIAO NMR calculations, combined with 2D NMR interpretation were used to complete the determination of their relative structures, which was in agreement with oxygenation pattern, and the unsaturation degree within the alkenyl chain were deduced from the predicted PKS domain arrangement. Targeted analysis allowed to map the abundance of xylasporins across termite-associated Xylaria isolates. While derivatives featuring a pentadienyl chain at C-8 were predominately produced by Xylaria sp. X187, Xylaria sp. X802 was found to produce only derivatives featuring a pentenyl chain at C-8, and thus differentiates those from cytosporins, which are characterized by a heptenyl chain and are produced by other fungal lineages. The weak antimicrobial inhibitory activity of only few of the isolated secondary metabolites supported our previous conclusion that termite-associated Xylaria might have adapted to such comb-specific stressors by reducing and specializing the secondary metabolome to minimize triggers that could stimulate alarm responses of the fungal mutualist and termites. Our combined metabolomic, structural analytical, and genomic study now allows to more accurately map the abundance of the PKS-terpenoid derived cytosporin/xylasporin across fungal genera and predict the structural features and relative stereochemistry based on our extensive GIAO NMR calculations encompassing calculated NMR data of more than 50 stereoisomers.
Method Section
General experimental procedures
Chemicals
All media, solvents, and, chemicals were purchased as follows: methanol, dichloromethane, ethyl acetate, acetonitrile (Th. Geyer, Renningen); water for analytical and preparative HPLC (Millipore, Germany), formic acid (Carl Roth, Germany), MeOH-d4, CDCl3 (Carl Roth, Germany), media ingredients (Carl Roth, Germany). Merck precoated silica gel F254 plates and RP-18 F254s plates were used for thin layer chromatography (TLC). Spots were detected on TLC under UV light or by heating after spraying with anisaldehyde-sulfuric acid.
Strains and culture conditions
A pre-culture of Xylaria sp. X8025,11,14 was prepared by inoculating 200 mL PDB with 1–2 agar cubes of a 2-week old Xylaria sp. X802 culture and incubated for 2 weeks at 30 °C and 150 rpm. Afterwards, the pre-culture was used to inoculate 20 PDA plates (92 × 16 mm), petri dishes filled with glass beads containing 10 mL liquid PDB and 10 mL rice-sawdust medium mixed in a 1:1 (v/v) ratio. The cultures incubated for 3 weeks at RT. Media served as negative controls. The cultures from liquid PDB and rice-sawdust medium were extracted with each 50 mL ethyl acetate overnight. Residues were combined, evaporated under reduced pressure, dissolved in MeOH and analysed using UPLC-ESI-HRMS.
Co-cultivation studies
Xylaria sp. X802 and Termitoymces sp. T153 were co-cultivated on PDA plate for three weeks as inoculum10,11. The mycelium of strain T153 was scratched from the surface of PDA plate and collected in sterile PBS (20 mL) to make nearly homogeneous mycelium suspension by vortex. Due to the different growth rate of both fungal strains, two different co-culture growth conditions on standard petri dish (9 cm diameter) containing 20 mL of 1/3 PDA medium were set-up. Method A: Xylaria sp. X802 and Termitoymces sp. T153 were inoculated simultaneously: one mycelium containing agar plug of X802 (1 × 1 cm) was placed in the middle of petri dish. Then 200 µL of T153 mycelium suspension was placed on the empty space of the same petri discs and evenly distributed by gentle spreading. Method B: Xylaria sp. X802 was grown for one week until vegetative mycelium of Xylaria sp. X802 was visible. Then Termitoymces sp. T153 mycelium suspension was introduced close to the X802 colony. Axenic cultures of Xylaria sp. X802 and Termitoymces sp. T153 were cultivated for the same time period as controls. To analyze the secreted metabolites within the zone of inhibition, the interaction zone from two-week co-cultivation plate was separated, cut into small pieces and extracted by 20 mL of dichloromethane (DCM) overnight. Similarly, mycelium of Xylaria sp. X802 and Termitoymces sp. T153 were separated, cut into small pieces and extracted by 20 mL of DCM for overnight respectively. Controls (Xylaria sp. X802 and Termitomyces sp. T153) were extracted in the same way. The DCM extract were collected by filtration and concentrated under reduced pressure using a SpeedVac. The resultant DCM extract were dissolved into LCMS grade MeCN as 0.1 mg/mL and submitted to UPLC-ESI-HRMS analysis.
HR-MS
High resolution ultra-high performance liquid chromatography electrospray ionization mass spectrometry (UPLC-ESI-HRMS) measurements were performed on a Dionex Ultimate3000 system coupled with a Luna Omega C18 column (100 × 2.1 mm, particle size 1.6 µm, pore diameter 100 Å, Phenomenex) combined with Q-Exactive Plus mass spectrometer (Thermo Scientific) equipped with an electrospray ion (HESI) source. Column oven was set to 40 °C; scan range of full MS was set to m/z 150 to 2000 with resolution of 70,000 and AGC target 3e6 and maximum IT 100 ms under positive and negative mode with centroid data type. MS2 was performed to choose top10 intensive ions under positive mode with resolution of 17,500 and AGC target 1e5 and maximum IT 50 ms and (N)CE (20/30/40) with centroid data type. The spray voltage (+) was set to 4000 volt, and (–) was set to 3300 volt. The capillary temperature (±) was set to 340 °C and probe heater temperature (±) was set to 200 °C. The sheath gas flow (±) was set to 35 L/min and Aux gas flow (±) to 5 L/min. Max spray current (+) and (–) was set to 100 volt. S-Lens RF level was set to 50.
GNPS analysis
Culture extracts were submitted to LC-ESI-HRMS metabolomics analysis under standard condition. Metabolites were separated under the gradient: 0–0.5 min, 5% B; 0.5–18 min, 5%–97% B; 18–23 min, 97% B; 23–25 min, 97%–5% B; 25–30 min, 5% B (A: 0.1% FA; B: MeCN with 0.1% FA), with flow rate of 0.3 mL/min and injection volume is 5 µL. Metabolomics raw data acquired on a Thermo QExactive Plus mass spectrometer was converted to 32-bit mzXML files using MSConvert GUI (ProteoWizard), in order to generate a mass spectral molecular networking using the global natural products social molecular networking platform (GNPS, https://gnps.ucsd.edu)6. Data analysis used default parameters, except for the cosine threshold, set to 0.7, minimum matched fragment ions of 4, network TopK 10, and for the tolerances of the precursor- and fragment ion masses, both set to 0.02 Da. The mass spectral network was assembled and visualized using Cytoscape35.
Analytical and preparative HPLC
High-performance liquid chromatography (HPLC) were performed on a Waters 1525 Binary HPLC pump with a Waters 996 Photodiode Array Detector (Waters Corporation, Milford, CT, USA). Semi-preparative HPLC used a Shimadzu Prominence HPLC System with SPD-20A/20AV Series Prominence HPLC UV-Vis Detectors (Shimadzu, Tokyo, Japan).
NMR
NMR spectra, including 1H-1H COSY, HSQC, HMBC, and ROESY experiments, were carried out using a Varian UNITY INOVA 800 NMR spectrometer operating at 800 MHz (1H) and 200 MHz (13C) or a Bruker Ascend 700 at 700 MHz (1H) and 175 MHz (13C), with chemical shifts given in ppm (δ). MestReNova ver. 12.0.1 was used to process the data for the NMR spectra.
Analytical characterization
Experimental ECD spectra in MeOH were acquired in a quartz cuvette of 1 mm optical path length on a JASCO J-1500 spectropolarimeter (Tokyo, Japan). IR spectra were acquired on a Bruker IFS-66/S FT-IR spectrometer. Optical rotations were obtained utilizing a Jasco P-1020 polarimeter (Jasco, Easton, MD, USA).
Crystallography data
X-ray analysis
The data set was collected using a Bruker D8 Venture diffractometer with a microfocus sealed tube and a Photon II detector. Monochromated MoKα radiation (λ = 0.71073) was used. Data were collected at 150(2) K and corrected for absorption effects using the multi-scan method. The structure was solved by direct methods using SHELXT36 and was refined by full matrix least squares calculations on F2 SHELXL201837 in the graphical user interface Shelxle38.
Refinement
All non H-atoms were located in the electron density maps and refined anisotropically. C-bound H atoms were placed in positions of optimized geometry and treated as riding atoms. Their isotropic displacement parameters were coupled to the corresponding carrier atoms by a factor of 1.2 (CH, CH2) or 1.5 (CH3). The O3 and N2 bonded H-atoms H3O and H2N were located in the electron density maps. Their positional parameters were refined using isotropic displacement parameters which were set at 1.2 (N2) and 1.5 (O3) times the Ueq value of the parent atoms. Restraints of 0.88(0.01) Å and 0.84(0.01) Å were used for the N-H and O-H bond lengths, respectively.
Crystal data for 9: C23H32N2O5, Mr = 416.50 gmol-1, size 0.200 × 0.060 × 0.060 mm3, orthorhombic, space group P 21 21 21, a = 9.2915(3), b = 10.7976(3), c = 22.0427(6) Å, α = 90, β = 90, γ = 90°, V = 2211.45(11) Å3, T = 150 K, Z = 4, ρcalcd. = 1.251 gcm-3, μ = 0.088 mm-1, multi-scan, transmin: 0.706, transmax: 0.746, F(000) = 896, 72893 reflections in h(-11/11), k(-13/13), l(-28/28), measured in the range 2.100° ≤ Θ ≤ 27.114°, completeness Θmax = 100%, 4888 independent reflections, Rint = 0.0521, 279 parameters, 2 restraints, R1obs = 0.0291, wR2obs = 0.0695, R1all = 0.0318, wR2all = 0.0718, GOOF = 1.055, Flack-parameter -0.0(2), largest difference peak and hole: 0.165 / -0.170 e Å-3.
Crystallographic data can be found in (Table S24) and has been deposited at the Cambridge Crystallographic Data Centre under CCDC-2347670 (Supplementary Data 2). This data can be obtained free of charge via www.ccdc.cam.ac.uk/conts/retrieving.html (or from the Cambridge Crystallographic Data Centre, 12, Union Road, Cambridge CB2 1EZ, UK; fax: (+44) 1223-336-033; or deposit@ccdc.cam.ac.uk).
Isolation of compounds 1-7 from Xylaria sp. X802
A flask with 50 mL PDB was inoculated with 2 agar cubes of a 2-week old Xylaria sp. X802 culture. The culture was incubated for 2 weeks at 30 °C and 150 rpm. Afterwards, the pre-culture was used to inoculate 120 PDA plates (20 mL PDA, 92 × 16 mm), which were kept at 26 °C for 14 days in the dark. Agar plates were then cut into pieces, soaked in EtOAc overnight. The organic extract was filtered of and the agar pieces were subsequently soaked in MeOH overnight, and filtrated again. The organic extracts were combined and dried under vacuo. The fungus crude extract (9.5 g) was suspended in distilled water (700 mL) and successively solvent-partitioned with EtOAc (700 mL) three times, yielding 2.0 g of residue. The EtOAc-soluble fraction was loaded onto silica-gel column chromatography and fractionated using a gradient solvent system of CH2Cl2-MeOH (100:1–1:1, v/v) to give five fractions (A-E). The fraction B (479 mg) was fractionated by preparative reversed-phase HPLC (Phenomenex Luna C18, 250 × 21.2 mm i.d., 5 μm) using CH3CN-H2O (1:9–1:0, v/v, gradient system, flow rate: 5 mL/min) to provide six subfractions (B1–B6). Subfraction B4 (56 mg) was isolated by a semi-preparative reversed-phase HPLC (Phenomenex Luna C18, 250 × 10.0 mm i.d., 5 μm) with 70% MeOH/H2O (isocratic system, flow rate: 2 mL/min) to yield compound 7 (1.7 mg, tR = 47.0 min). The fraction C (160 mg) was separated by preparative reversed-phase HPLC (Phenomenex Luna C18, 250 × 21.2 mm i.d., 5 μm) using CH3CN-H2O (1:9–1:0, v/v, gradient system, flow rate: 5 mL/min) to yield five subfractions (C1–C5). Compound 1 (1.4 mg, tR = 24.0 min) was isolated from subfraction C3 (40 mg) by a semi-preparative reversed-phase HPLC (Phenomenex Luna C18, 250 × 10.0 mm i.d., 5 μm) with 40% MeOH/H2O (isocratic system, flow rate: 2 mL/min). Fraction C4 (20 mg) was purified by a semi-preparative reversed-phase HPLC (Phenomenex Luna C18, 250 × 10.0 mm i.d., 5 μm) with 45% MeOH/H2O (isocratic system, flow rate: 2 mL/min) to afford compound 2 (2.2 mg, tR = 30.0 min). The fraction D (101 mg) was subjected onto a preparative reversed-phase HPLC (Phenomenex Luna C18, 250 × 21.2 mm i.d., 5 μm) and separated by CH3CN-H2O (2:8–1:0, v/v, gradient system, flow rate: 5 mL/min) to yield five subfractions (D1–D5). Subfraction D2 (18 mg) was isolated by a semi-preparative reversed-phase HPLC (Phenomenex Luna C18, 250 × 10.0 mm i.d., 5 μm) utilizing 60% MeOH/H2O (isocratic system, flow rate: 2 mL/min) to give compounds 4 (1.2 mg, tR = 31.0 min), 3 (1.5 mg, tR = 39.0 min), 6 (1.8 mg, tR = 45.0 min), and 5 (2.7 mg, tR = 48.0 min).
Isolation of compounds 8 and 9 from Xylaria sp. X187
A culture was prepared by inoculating 50 mL PDB with 2 agar cubes of a 2-week old Xylaria sp. X187 culture. The culture was incubated for 2 weeks at 30 °C and 150 rpm. Afterwards, the pre-culture was used to inoculate 20 PDA plates (150 × 20 mm), which were kept at 26 °C for one month in the dark. Agar plates were then cut into pieces, soaked in EtOAc overnight. The crude extract (720 mg) was loaded onto C18 SPE cartridge and fractionated using a gradient solvent system of H2O-CH3CN (100:0-0:100, v/v) to give seven fractions (A-F). Xylasporin I (8) (1.2 mg, tR = 16.4 min) was isolated from fraction B H2O-CH3CN (80:20, v/v) by semi-preparative reversed-phase HPLC (Phenomenex Luna C18, 250 × 10.0 mm i.d., 5 μm) with CH3CN-H2O (1:9–3:7, v/v, gradient system, flow rate: 3 mL/min). The fraction D was subjected onto a semi-preparative reversed-phase HPLC (Phenomenex Luna C18, 250 × 10.0 mm i.d., 5 μm) and separated by CH3CN-H2O (2:8–5:5, v/v, gradient system, flow rate: 5 mL/min) to yield xylacremolide B (9) (5 mg, tR = 27.9 min).
Physical chemical data
Xylasporin A (1): Pale yellow oil; [α]25D 15.1 (c 0.09, MeOH); IR (KBr) νmax 3379, 2921, 1786, 1652, 1048 cm−1; ECD (MeOH) λmax (△ε) 228 (1.7), 265 (4.1) nm; 1H (800 MHz) and 13C NMR (200 MHz), see Tables S1 and S2, respectively; positive HR-ESI-MS m/z 709.3430 [2 M + H]+ (Calcd. for C36H53O14, 709.3435).
Xylasporin B (2): Pale yellow oil; [α]25D -7.4 (c 0.05, MeOH); IR (KBr) νmax 3422, 2919, 1784, 1552, 1248 cm−1; ECD (MeOH) λmax (△ε) 245 (3.7) nm; 1H (800 MHz) and 13C NMR (200 MHz), see Tables S1 and S2, respectively; positive HR-ESI-MS m/z 293.1742 [M-H2O + H]+ (Calcd. for C17H25O4, 293.1753).
Xylasporin C (3): Pale yellow oil; [α]25D -11.2 (c 0.08, MeOH); IR (KBr) νmax 3412, 2903, 1779, 1498, 1228 cm−1; ECD (MeOH) λmax (△ε) 242 (3.3) nm; 1H (800 MHz) and 13C NMR (200 MHz), see Tables S1 and S2, respectively; positive HR-ESI-MS m/z 621.3627 [2 M + H]+ (Calcd. for C34H53O10, 621.3639).
Xylasporin D (4): Pale yellow oil; [α]25D 8.7 (c 0.06, MeOH); IR (KBr) νmax 3405, 2895, 1770, 1454, 1213 cm−1; ECD (MeOH) λmax (△ε) 251 (3.2) nm; 1H (800 MHz) and 13C NMR (200 MHz), see Tables S1 and S2, respectively; positive HR-ESI-MS m/z 351.1784 [M+Na]+ (Calcd. for C17H28O6Na, 351.1784).
Xylasporin E (5): Pale yellow oil; [α]25D 5.3 (c 0.04, MeOH); IR (KBr) νmax 3412, 2903, 1787, 1464, 1223 cm−1; ECD (MeOH) λmax (△ε) 245 (3.6) nm; 1H (800 MHz) and 13C NMR (200 MHz), see Tables S1 and S2, respectively; positive HR-ESI-MS m/z 657.3850 [2 M + H]+ (Calcd. for C34H57O12, 657.3850).
Xylasporin F (6): Pale yellow oil; [α]25D 8.1 (c 0.05, MeOH); IR (KBr) νmax 3402, 2895, 1780, 1458, 1215 cm−1; ECD (MeOH) λmax (△ε) 245 (3.4) nm; 1H (800 MHz) and 13C NMR (200 MHz), see Tables S1 and S2, respectively; positive HR-ESI-MS m/z 657.3855 [2 M + H]+ (Calcd. for C34H57O12, 657.3850).
Xylasporin I (8): White powder; 1H (700 MHz) and 13C NMR (150 MHz), see Table S20; positive HR-ESI-MS m/z 309.1694 [M + H]+ (Calcd. for C17H25O5, 309.1697).
Xylacremolide B (9): White crystals; 1H (700 MHz) and 13C NMR (150 MHz), see Table S23; positive HR-ESI-MS m/z 417.2369 [M + H]+ (Calcd. for C23H33N2O5, 417.2384).
Conformational search and geometry optimization
All conformers were acquired through Marvin module (ChemAxon). Minimize molecular conformations were obtained using an industry standard MMFF94 force field and a generic Dreiding force field. Conformers proposed in this study within 5 kJ/mol found in the MMFF force field were selected for geometry optimization by Gaussian 16 program package39 at B3LYP/6-31 + G(d,p) level. The solvent (MeOH or CDCl3) effect was considered in the PCM approximation (Supplementary Data 1).
Chemical shifts calculation and DP4+ analysis
Geometrically optimized conformers for possible diastereomers of 1–6 and 8 were proceeded to calculation of gauge-invariant atomic orbital (GIAO) magnetic shielding tensors at the B3-LYP/6-31 + G(d,p) level using Gaussian 16 program package39. Chemical shift values were calculated from the magnetic shielding tensors using equation 1 where \(\delta\) is the calculated NMR chemical shift for nucleus \(x\), and\(\,{\sigma }^{o}\) is the shielding tensor for the proton and carbon nuclei in tetramethylsilane calculated with the B3-LYP/6-31 + G(d,p) basis set (Supplementary Data 1).
The calculated unscaled NMR properties of the optimized structures were averaged based on the Boltzmann populations of conformers and scaled chemical shifts values were obtained using equation 2.
Each experimental set of data was separately compared in detail with calculated ones, specifically for each accounted atom. Experimental and calculated chemical shits (δ) were compared using the Δδ parameter according equation 3.
where δcalc and δexp are the calculated and experimental chemical shift values, respectively.
After calculating all Δδ values, mean absolute error (MAE) values and DP4+ probabilities were computed for determining which calculated set of data fits better with the experimental one. The MAE is defined as the summation (∑) of the n computed absolute δ error values (Δδ) normalized to the number of Δδ errors considered (n) (equation 4).
DP4+ probability analysis was processed upon using an Excel sheet from ref. 19.
ECD spectra calculation
ECD calculations of 1 conformers were performed at the identical theory level and basis sets with Gaussian 16 program package39. Calculated ECD spectra were simulated by overlying each transition, where σ is the width of the band at the height 1/e (equation 5). Ei and Ri are the excitation energies and rotatory strengths for transition i, respectively. In the present study, the value of σ was 0.30 eV. The excitation energies and rotational strengths for ECD spectra were calculated, and ECD visualization was performed using GaussView 6.0.
Antimicrobial activity assays. A
Antibacterial aactivity assay was performed according to the NCCLS (National Committee for Clinical Laboratory Standards) with compound (1.0 mg/mL in DMSO) and ciprofloxacin (5 µg/mL in aqua dest. (cip.) as control. Bacterial test strains included Bacillus subtilis sp. 6633, Staphylococcus aureus SG511, E.coli sp. SG458, Pseudomonas aeruginosa sp. SG137, MRSA Staphylococcus aureus sp. 134/94, VRSA Enterococcus faecalis, Mycobacterium vaccae sp. 10670. The antifungal activity assays were performed according to ref. 40. B) Aspergillus nidulans RMS011 was chosen as a test strain for antibacterial testing. As a reference the antifungal agent carboxine (AppliChem GmbH, Germany) was used. Stock solutions (1 mg/mL in MeOH) of the tested substances were added in 96 well plates and MeOH was evaporated at room temperature. For inoculation 150 µL of conidia suspension in aspergillus minimal medium (AMM, supplemented with 3 mg/L p-aminobenzoic acid and 5 mM L-arginine) and resazurin (0.002% (w/v); Sigma-Aldrich Chemie GmbH, Germany) were used. Conidia concentration was 2.5 × 105 conidia/mL. Assays were incubated at 37 °C for 20 h in a Synergy H1 plate reader (BioTek Instruments GmbH, Germany) and fluorescence was determined at the wavelengths 570 nm (excitation) and 615 nm (emission). The growth inhibition was calculated by normalization to the positive control without supplementation of substances and the negative control without inoculum. All assays were carried out in triplicates. Shown is the relative inhibition (in %) of epoxycytochalasine Q (Ecc), pseudoxylallemycin A (Pam), cytosporins N-R (2-6), 1-hydroxy-2-hydroxymethyl-3-pent-1-enylbenzene (7) and SPE fraction eluted with MeOH of Xylaria sp. X802 raw extract (SPE) on A. nidulans RMS011 growth after 20 h. The percentage of inhibition equal to 0 is considered normal growth and equal to 100 is complete growth inhibition. Growth control (Control) was set up without supplementation of compounds. The antifungal agent carboxine (Cbx) was used as a reference. All of the experiments were performed in triplicate and the standard deviations are shown (Supplementary Fig. S62).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Supplementary Information: contains copies of GNPS figures, all NMR spectra, HRMS spectra of clean compounds, comparison of calculated and measured NMR data, DP4+ probability analyses, BLAST results of antimicrobial activity screening, and in silico analysis of the px gene cluster within the genome of Xylaria sp., and crystal data and structure refinement data for compound 9. Supplementary Data 1: contains lists of calculated 1H and 13C chemical shifts for each determined conformer. Supplementary Data 2: Crystallographic Information File (CIF) for structure of compound 9. The X-ray crystallographic coordinates for structure of compound 9 reported in this study have also been deposited at the Cambridge Crystallographic Data Centre (CCDC), under deposition numbers CCDC-2347670. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data_request/cif. All data generated or analyzed during this study are included in this published article and its supplementary files. NMR data files and supplementary mass-spectrometry data files have also been deposited at Zenodo under the https://doi.org/10.5281/zenodo.10978114.
References
Spatafora, J. W. et al. The fungal tree of life: from molecular systematics to genome-scale phylogenies. Microbiol. Spectr. 5, https://doi.org/10.1128/microbiolspec.funk-0053-2016 (2017).
Becker, K. & Stadler, M. Recent progress in biodiversity research on the Xylariales and their secondary metabolism. J. Antibiot. 74, 1–23 (2021).
Rokas, A., Mead, M. E., Steenwyk, J. L., Raja, H. A. & Oberlies, N. H. Biosynthetic gene clusters and the evolution of fungal chemodiversity. Nat. Prod. Rep. 37, 868–878 (2020).
Kuhnert, E. et al. Secondary metabolite biosynthetic diversity in the fungal family Hypoxylaceae and Xylaria hypoxylon. Stud. Mycol. 99, 100118 (2021).
Visser, A. A. et al. Levels of specificity of Xylaria species associated with fungus-growing termites: a phylogenetic approach. Mol. Ecol. 18, 553–567 (2009).
Hsieh, H. M. et al. Phylogenetic status of Xylaria subgenus Pseudoxylaria among taxa of the subfamily Xylarioideae (Xylariaceae) and phylogeny of the taxa involved in the subfamily. Mol. Phylogenet Evol. 54, 957–969 (2010).
Li, H., Young, S. E., Poulsen, M. & Currie, C. R. Symbiont-mediated digestion of plant biomass in fungus-farming insects. Annu Rev. Entomol. 66, 297–316 (2021).
Visser, A. A., Kooij, P. W., Debets, A. J. M., Kuyper, T. W. & Aanen, D. K. Pseudoxylaria as stowaway of the fungus-growing termite nest: Interaction asymmetry between Pseudoxylaria, Termitomyces and free-living relatives. Fung. Ecol. 4, 322–332 (2021).
Wisselink, M. & Aanen, D. K. van’t Padje A. The longevity of colonies of fungus-growing termites and the stability of the symbiosis. Insects 11, 527 (2020).
Fricke, J. et al. Adaptations of Pseudoxylaria towards a comb-associated lifestyle in fungus-farming termite colonies. ISME J. 17, 733–747 (2023).
Schmidt, S., Kildgaard, S., Guo, H., Beemelmanns, C. & Poulsen, M. The chemical ecology of the fungus-farming termite symbiosis. Nat. Prod. Rep. 39, 231–248 (2022).
Guo, H. et al. Cyclic tetrapeptides with rare allenyl modifications isolated from Pseudoxylaria sp. X802: a competitor of fungus-growing termite cultivars. Org. Lett. 18, 3338–3341 (2016).
Schalk, F. et al. GNPS-guided discovery of xylacremolide C and D, evaluation of their putative biosynthetic origin and bioactivity studies of xylacremolide A and B. RSC Adv. 11, 18748–18756 (2021).
Schalk, F. et al. Targeted discovery of tetrapeptides and cyclic polyketide-peptide hybrids from a fungal antagonist of farming termites. Chembiochem 21, 2991–2996 (2020).
Adnani, N., Rajski, S. R. & Bugni, T. S. Symbiosis-inspired approaches to antibiotic discovery. Nat. Prod. Rep. 34, 784–814 (2017).
Keller, N. P. Fungal secondary metabolism: regulation, function and drug discovery. Nat. Rev. Microbiol. 17, 167–180 (2019).
Wang, M. et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 34, 828–837 (2016).
Ciavatta, M. L. et al. Cytosporin-related compounds from the marine-derived fungus Eutypella scoparia. Tetrahedron 64, 5365–5369 (2008).
Grimblat, N., Zanardi, M. M. & Sarotti, A. M. Beyond DP4: an Improved probability for the stereochemical assignment of isomeric compounds using quantum chemical calculations of NMR Shifts. J. Org. Chem. 80, 12526–12534 (2015).
Stevens-Miles, S. et al. Discovery of an angiotensin II binding inhibitor from a Cytospora sp. using semi-automated screening procedures. J. Antibiot. 49, 119–123 (1996).
Liu, Z., Jensen, P. R. & Fenical, W. A cyclic carbonate and related polyketides from a marine-derived fungus of the genus. Phoma. Phytochem. 64, 571–574 (2003).
Mehta, G. & Roy, S. Enantioselective total synthesis of (+)-eupenoxide and (+)-phomoxide: revision of structures and assignment of absolute configuration. Org. Lett. 6, 2389–2392 (2004).
Akone, S. H. et al. new epoxyquinols from the endophytic fungus Pestalotiopsis theae. Tetrahedron Lett. 54, 6751–6754 (2013).
Rukachaisirikul, V. et al. γ-Butyrolactone, cytochalasin, cyclic carbonate, eutypinic acid, and phenalenone derivatives from the soil fungus Aspergillus sp. PSU-RSPG185. J. Nat. Prod. 77, 2375–2382 (2014).
Liao, H. X., Sun, D. W., Zheng, C. J. & Wang, C. Y. A new hexahydrobenzopyran derivative from the gorgonian-derived fungus Eutypella sp. Nat. Prod. Res. 31, 1640–1646 (2017).
Zhang, Y. X. et al. meroterpenoids from the arctic fungus Eutypella sp. D-1. J. Nat. Prod. 82, 3089–3095 (2019).
Rivera-Chávez, J. et al. Cuautepestalorin, a 7,8-Dihydrochromene-Oxoisochromane Adduct Bearing a Hexacyclic Scaffold from Pestalotiopsis sp. IQ-011. Org. Lett. 21, 3558–3562 (2019).
Yu, X. et al. Polyketide derivatives from mangrove derived endophytic fungus Pseudopestalotiopsis theae. Mar. Drugs 18, 129 (2020).
Zhang, Y. H. et al. Anti-inflammatory polyketides from the marine-derived fungus Eutypella scoparia. Mar. Drugs 20, 486 (2022).
Davis, R. A. et al. from an endophytic fungus of the genus. Eupenicillium. Phytochem. 66, 2771–2775 (2005).
Nies, J., Ran, H., Wohlgemuth, V., Yin, W. B. & Li, S. M. Biosynthesis of the prenylated salicylaldehyde flavoglaucin requires temporary reduction to salicyl alcohol for decoration before reoxidation to the final product. Org. Lett. 22, 2256–2260 (2020).
Terlouw, B. R. et al. MIBiG 3.0: a community-driven effort to annotate experimentally validated biosynthetic gene clusters. Nucleic Acids Res. 51, D603–D610 (2023).
Liu, L., Tang, M. C. & Tang, Y. Fungal highly reducing polyketide synthases biosynthesize salicylaldehydes that are precursors to epoxycyclohexenol natural products. J. Am. Chem. Soc. 141, 19538–19541 (2019).
Zhao, Z., Ying, Y., Hung, Y. S. & Tang, Y. Genome mining reveals Neurospora crassa can produce the salicylaldehyde sordarial. J. Nat. Prod. 82, 1029–1033 (2019).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Sheldrick, G. M. SHELXT – Integrated space-group and crystal-structure determination. Acta Cryst. A 71, 3–8 (2015).
Sheldrick, G. M. Crystal structure refinement with SHELXL. Acta Cryst. C. 71, 3–8 (2015).
Hübschle, C. B., Sheldrick, G. M. & Dittrich, B. ShelXle: a Qt graphical user interface for SHELXL. J. Appl. Cryst. 44, 1281–1284 (2011).
Frisch, M. J. et al. Gaussian 16 Rev. B.01 (Gaussian Inc., 2016).
Monteiro, M. C. et al. A new approach to drug discovery: high-throughput screening of microbial natural extracts against Aspergillus fumigatus using resazurin. J. Biomol. Screen. 17, 542–549 (2012).
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2019R1A5A2027340 and 2021R1A2C2007937) and National Research Foundation of Korea (NRF) funded by the Ministry of Education (2020R1A6A3A03037782). This work was also supported under the framework of international cooperation program managed by the National Research Foundation of Korea (2020K2A9A2A06037042). This study was funded by the German Research Foundation (DFG, Deutsche Forschungsgemeinschaft) under Project-ID 239748522 – CRC 1127 (project A6), project BE 4799/4-1 under Project-ID 438841444, and the Germany’s Excellence Strategy under Project-ID 390713860 - EXC 2051. We thank Mrs. Heike Heinecke (Hans Knöll Institute) for recording NMR spectra and Mrs. Andrea Perner (Hans Knöll Institute) for HRMS measurements. We thank Dr. Fabien Fontaine-Vive for launching DFT and TDDFT calculations as well as Université Côte d’Azur and OPAL infrastructure for providing Azzurra High-Performance Computing. Instrumentation and technical assistance for crystallographic data were provided by the Service Center X-ray Diffraction, with financial support from Saarland University and German Science Foundation project number INST 256/506-1 (D8 Venture).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
S.R.L., M.D., J.F., H.G., F.S., S.B., J.S.Y., B.S., C.B. designed research; S.R.L., M.D., J.F., H.G., S.B., F.S., S.B., J.S.Y., performed the experiments; S.R.L., M.D., J.F., H.G., F.S., J.S.Y., B.M., S.Y.J., C.B., K.H.K. analysed data; B.S., K.H.K., C.B. supervised, provided funds and infrastructure. S.R.L., M.D., J.F., H.G., F.S., B.M., J.S.Y., C.B. generated the figures and S.R.L., M.D., J.F., B.S., K.H.K., C.B. wrote the manuscript with input from all authors
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests. H.G. is a Senior Editor for Communications Chemistry, but was not involved in the editorial review of, or the decision to publish this article.
Peer review
Peer review information
Communications Chemistry thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lee, S.R., Dayras, M., Fricke, J. et al. Molecular networking and computational NMR analyses uncover six polyketide-terpene hybrids from termite-associated Xylaria isolates. Commun Chem 7, 129 (2024). https://doi.org/10.1038/s42004-024-01210-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42004-024-01210-6
- Springer Nature Limited