Abstract
Covalent attachment of methoxy poly(ethylene) glycol (mPEG) to therapeutic molecules is widely employed to improve their systemic circulation time and therapeutic efficacy. mPEG, however, can induce anti-PEG antibodies that negatively impact drug therapeutic effects. However, the underlying mechanism for specific binding of antibodies to mPEG remains unclear. Here, we determined the first co-crystal structure of the humanized 15-2b anti-mPEG antibody in complex with mPEG, which possesses a deep pocket in the antigen-binding site to accommodate the mPEG polymer. Structural and mutational analyses revealed that mPEG binds to h15-2b via Van der Waals and hydrogen bond interactions, whereas the methoxy group of mPEG is stabilized in a hydrophobic environment between the VH:VL interface. Replacement of the heavy chain hydrophobic V37 residue with a neutral polar serine or threonine residue offers additional hydrogen bond interactions with methoxyl and hydroxyl groups, resulting in cross-reactivity to mPEG and OH-PEG. Our findings provide insights into understanding mPEG-binding specificity and antigenicity of anti-mPEG antibodies.
Similar content being viewed by others
Introduction
PEGylation is a widely applied method that covalently conjugates therapeutics with methoxy polyethylene glycol (mPEG) for improving their pharmaceutical and pharmacokinetic properties1,2,3. mPEGylated therapeutics have been approved by the U.S. Food and Drug Administration, including small molecular drugs4,5, proteins6,7,8,9 and nanoparticles10,11. For example, low molecular weight mPEG can be conjugated with hydrophobic small molecular drugs4,5 to improve their water solubility and decrease systemic toxicity4,5. On the other hand, high molecular weight mPEG is often attached to the surface of therapeutic proteins to prolong their serum half-life and protect against proteolytic degradation7,8,9. Incorporation of mPEG molecules on nanoparticles such as Doxil (mPEG-liposomal doxorubicin) and COVID-19 mRNA vaccines (mPEG-containing lipid nanoparticles-mRNA) can reduce unwanted uptake by the reticuloendothelial system in vivo10 and prevent lipid nanoparticles-mRNA aggregation during storage as an aqueous dispersion12, respectively.
mPEG is a biocompatible, well-tolerated polymer but many preclinical and clinical studies report that mPEGylated therapeutic molecules can trigger anti-PEG antibody production leading to reduced therapeutic efficacy13,14,15. For instance, anti-PEG antibodies can form immune complexes with mPEGylated therapeutics to induce complement activation resulting in accelerated blood clearance (ABC) via uptake into macrophages in the liver15,16. In addition, drug encapsulated mPEGylated liposomes can be destabilized by anti-PEG antibody-mediated complement activation, resulting in rapid drug leakage and diminished anti-tumor activity14,17. Surprisingly, pre-existing anti-PEG antibodies have been discovered in healthy donors18,19, raising concerns for induction of severe allergic reactions in some individuals who receive mPEGylated therapeutics, including PEGylated protein drugs and COVID-19 mRNA vaccines20,21.
Anti-PEG antibodies can be classified into two broad groups depending on their binding specificity. Antibodies that bind to the repeating ethylene oxide repeats of PEG are termed “backbone-specific”, whereas antibodies that require the terminal methoxy group for binding are termed “methoxy-specific”18,22,23. Previous studies have shown that backbone-specific anti-PEG antibodies can negatively impact the biodistribution and therapeutic efficacy of mPEGylated medicines13,14. The crystal structures of two backbone-specific anti-PEG monoclonal antibodies (3.3 and 6.3) revealed that dimerization of these antibodies is essential for PEG-binding24,25. Although PEG used in most therapeutic medicines is terminated with a methoxy group, the clinical relevance of methoxy-specific anti-mPEG antibodies remains largely unknown. Previous studies demonstrated that foreign proteins modified with mPEG induce higher titers of anti-PEG antibodies as compared to proteins modified with PEG in animals26,27, but mPEG-modified liposomes trigger reduced accelerated blood clearance as compared to PEG-modified liposomes28. The mechanism of binding of methoxy-specific anti-mPEG antibodies is also unknown.
In this study, we elucidate the binding mode of the humanized h15-2b antibody, which selectively binds to mPEG. We describe the crystal structure of h15-2b Fab in complex with mPEG. Structure-guided mutagenesis was performed to define crucial amino acid residues involved in binding to mPEG. Our findings provide new understandings of anti-PEG antibody specificity and mPEG antigenicity.
Results
h15-2b Fab specifically binds to mPEG via the terminal methoxy group
Anti-PEG antibodies can be classified as backbone-specific and methoxy-specific. For instance, humanized 6.3 (h6.3) binds to the repeating ethylene oxide repeats in the PEG backbone, whereas h15-2b binds to PEG terminated with a methoxy moiety18,22. To examine the binding specificity of these anti-PEG antibodies, PEG molecules with various end groups, including mPEG5K-NH2, OH-PEG5K-NH2, and SH-PEG3.5K-NH2, which contain a terminal methoxy, hydroxyl, or thiol groups, respectively, were immobilized on the surface of ELISA plates via the amine group18. Comparison of the binding specificity of recombinant Fab fragments derived from h6.3 or h15-2b as well as a negative control human anti-GFP Fab shows that h15-2b Fab strongly binds to mPEG5K-NH2 but not to OH-PEG5K-NH2 or SH-PEG3.5K-NH2 (Fig. 1), demonstrating that h15-2b displays selectivity for the terminal methoxy group of mPEG. By contrast, h6.3 Fab binds to all variant PEG molecules, indicating that h6.3 Fab binds to the repeating subunits of the PEG backbone (Fig. 1).
Microplate wells coated with amino-PEG molecules terminated (a) methoxy, (b) hydroxyl, or (c) thiol groups were incubated with graded concentrations of backbone-specific (h6.3, black circle) or methoxy-specific (h15-2b, blue open triangle) anti-PEG Fabs or anti-GFP Fab (negative control, open square). After washing, the Fab binding was determined by adding HRP-conjugated goat anti-human F(ab’)2 fragment specific antibodies, followed by ABTS substrate. The results show the mean absorbance values (405 nm) ± standard deviation (n = 3). The data are representative of three independent experiments.
h15-2b Fab complex with mPEG deep in the VH:VL interface
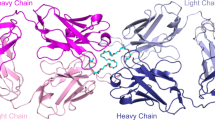
We performed X-ray crystallography to determine the three-dimensional structure of h15-2bmPEG complex. The h15-2bmPEG complex was crystallized in an orthorhombic space group P21 with the unit cell dimensions: a = 63.99 Å, b = 112.29 Å, c = 158.73 Å. The crystal structure of h15-2bmPEG was determined by the molecular replacement (MR) method using the structure of the anti-ErbB2 Fab2C4 (Protein Data bank entry 1L7I) as the search model29. The final model of h15-2bmPEG was refined to a resolution of 2.08 Å with an R factor of 21.1% and an Rfree of 24.5%. Data processing and refinement statistics are given in Table 1. The structure of h15-2bmPEG contains four copies of h15-2b Fab in an asymmetric unit, which all share nearly identical structures with small root-mean-square deviations (rmsds) of 0.161–0.417 Å for Cα atom superposition (Supplementary Fig. 1a; Supplementary Data 1). The light chains of the four copies of h15-2b Fab had varying electron density from the N- to C-termini, with the starting from the first residue and ending between residues 211 and 213. The heavy chains of the four copies of h15-2b Fab could be reliably traced between residues 1 and 214–216, with the region between residues 127 and 135 lacking significant density. Each of h15-2b Fabs shows that a hook-shaped mPEG fragment corresponding to 7 ethylene oxide subunits (about 300 Da) was bound into the pocket formed by the VH and VL domains of h15-2b Fab (Fig. 2a). The superposition of the four copies of h15-2b Fab reveals that the binding poses of the mPEG fragments are almost identical (Supplementary Fig. 1b). It forms a van der Waals network with aromatic side chains in Y32H, W33H, and Y102H from the heavy chain, and Y36L, W50L, and Y91L from the light chain (Fig. 2b). In addition, the PEG backbone wraps around R101H. The terminal methoxy group of the mPEG fragment is buried in the VH:VL interface pocket, where V37H, W104H, L89L, and F98L create a hydrophobic environment to accommodate the methoxy group (Fig. 2c). In this study, one of the h15-2b Fab structures (Chain A for light chain and Chain B for heavy chain in the atomic coordinates of h15-2bmPEG) was utilized for figure generation (Figs. 2–4).
a Overall crystal structure of the mPEG molecule and anti-mPEG h15-2b Fab complex (Light chain in green; Heavy chain in cyan). The mPEG fragment bound by h15-2b Fab is shown as a yellow stick model. Superposition of (b) mPEG backbone-specific binding residues and (c) methoxy-specific binding residues in the h15-2bmPEG crystal structure are colored (Light chain in green; Heavy chain in cyan) and displayed in a transparent mode. The methoxy group of mPEG is indicated with an arrow.
a CR-like structures of mPEG and anti-mPEG h15-2b Fab complex (Light chain in green; Heavy chain in cyan). The mPEG fragment bound by h15-2b Fab is shown as a yellow stick model. b–d Microplate wells coated with mPEG5K-NH2 were incubated with graded concentrations of parental h15-2b or relevant variants corresponding to CR-like structures of mPEG interaction. After 1 h, the wells were washed, and antibody binding was determined by adding HRP-conjugated goat anti-human F(ab’)2 fragment specific antibodies, followed by ABTS substrate. The results show the mean absorbance values (405 nm) ± standard deviation (n = 3). The data are representative of three independent experiments.
a Microplate wells coated with mPEG5K-NH2 and OH-PEG5K-NH2 were incubated with graded concentrations of parental h15-2b or relevant variants corresponding to methoxy group interacting residues. After 1 h, the wells were washed, and antibody binding was determined by adding HRP-conjugated goat anti-human F(ab’)2 fragment specific antibodies, followed by ABTS substrate. The results show the mean absorbance values (405 nm) ± standard deviation (n = 3). The data are representative of three independent experiments. b Simulated binding models of mPEG or OH-PEG (yellow sticks) interacting with the V37HT variant. Hydrogen bonds are indicated by red dashes.
It should be noted that the co-crystal structure of h15-2b Fab and mPEG was crystallized by using a 6:1 molar ratio of OH-PEG6K (15 mmol L−1) and mPEG2K (2.5 mmol L−1) as a cryoprotectant and an antigen, respectively. Although we demonstrated the specificity of h15-2b toward mPEG but not OH-PEG in Fig. 1, it raises a question of whether h15-2b Fab forms a complex with mPEG2K or more abundant OH-PEG6K during protein crystallization. To address this issue, we performed competitive ELISA with OH-PEG6K and mPEG2K. Microplates coated with mPEG2K-NH2 or TNF-α (2 µmol L−1) were competed with either OH-PEG6K or mPEG5K (12 µmol L−1) in the presence of a fixed amount of Fab. A detailed experimental process can be found in Supplementary Methods. We found that both OH-PEG6K and mPEG5K blocked the binding of 6.3 Fab (backbone-specific) to immobilized mPEG2K-NH2 in a dose-dependent manner (Supplementary Fig. 2a, b). By contrast, OH-PEG6K is a poor competitor, while mPEG5K is a strong competitor for binding of the h15-2b Fab (methoxy-specific) to immobilized mPEG2K-NH2. Therefore, the competitive ELISA results suggest that h15-2b Fab can specifically bind to mPEG and tolerate the presence of 6-fold molar excess of OH-PEG6K (Supplementary Fig. 2). Furthermore, neither OH-PEG6K nor mPEG5K affects the binding of an anti-TNF-α control Fab (Humira) to immobilized TNF-α, demonstrating the specificity of the competition reaction (Supplementary Fig. 2c, d). Taken together, h15-2b is more likely to form a complex with mPEG2K instead of OH-PEG6K.
Characterization of critical residues for h15-2b mPEG interaction
To verify critical amino acid residues that interact with the mPEG chain, we performed site-directed mutagenesis30 focusing on the amino acids around the backbone of mPEG. Previous studies suggest that aromatic residues play major roles in binding crown ether (CR)-like structure in the PEG backbone24,25. Since H-bonds also provide a significant intermolecular force for interaction, we investigated the importance of H-bonding interactions between aromatic side chains and the ether oxygen atoms of mPEG. Thus, we mutated Y32H, W33H, N35H, Y102H, Y36L, W50L, and Y91L into different aromatic residues (Y or W or F) that possess aromatic rings with or without the capability to form H-bonds.
Aromatic residues, including Y32H, W33H, Y102H, and W50L, clamp the first semicircular CR-like structure of mPEG (Fig. 3a). Mutation of Y32H and Y102H to phenylalanine (Y32HF and Y102HF) produce similar mPEG-binding activity as parental h15-2b, suggesting that the benzene ring side-chain is critical for mPEG interaction through hydrophobic forces but not through H-bonds (Fig. 3b; Supplementary Table 1). By contrast, Y32HW and Y102HW exhibited impaired mPEG-binding activity, indicating that the bulky indole group of tryptophan might negatively impact the mPEG interaction (Fig. 3b; Supplementary Table 1). Mutating W33H and W50L to tyrosine resulted in similar or minor decreased mPEG-binding activity, whereas W33HF and W50LF displayed a clear reduction of mPEG-binding, suggesting that the indole ring of W33H and W50L might participate in mPEG-binding via H-bonds (Fig. 3c; Supplementary Table 1). The second semicircular CR-like structure is clamped by W33H, N35H, Y36L, and Y91L (Fig. 3a). Both Y91LF and Y91LW showed similar mPEG-binding activity as the parental h15-2b, indicating that the aromatic side chain plays a major role to contact the mPEG chain without the need of H-bonding (Fig. 3d; Supplementary Table 1). Structural analysis also showed that the phenolic hydroxyl group of Y36L forms an H-bond with the ether oxygen atoms of mPEG. Indeed, mutation of Y36L to phenylalanine (Y36LF) completely abolished mPEG-binding activity, indicating an important role of Y36L for mPEG interaction (Fig. 3d; Supplementary Table 1). In addition, N35H also forms an H-bond with the ether oxygen atoms of mPEG. Thus, mutation of N35H to aromatic residues leads to the elimination of the mPEG-binding activity (Fig. 3d; Supplementary Table 1). These results suggest that the flexible mPEG chain is captured by h15-2b Fab through van der Waals forces from aromatic side chains that clamp the PEG backbone with possible hydrogen bond formation between the hydroxyl group or indole nitrogen atom of Y36L, W33 H, and W50L and oxygen atoms of mPEG. Furthermore, it has been reported that lysine and arginine residues can coordinate with PEG to form a CR-like structure, in which a cyclic PEG is wrapped as crown ether analogue24,31. The second semicircular CR-like structure is also wrapped around the guanidine group of R101H, which forms a possible H-bond with the oxygen atoms of mPEG (Fig. 3a). Unsurprisingly, mutation of R101H to alanine (R101HA) completely abolished mPEG-binding (Fig. 3d; Supplementary Table 1). We further generated a R101HK mutant to investigate whether lysine is able to interact with mPEG to form the CR-like structure. However, due to the crucial interaction between R101H and Y96L (Supplementary Fig. 3), the CR-like structure of mPEG mediated by R101HK mutant might be unstable, resulting in the elimination of the mPEG-binding (Fig. 3d; Supplementary Table 1). This result supports that the interaction between R101H and Y96L is important for mPEG binding and might stabilize the CR-like structure of mPEG.
Characterization of important residues of h15-2b for mPEG-binding
Based on the crystal structure, four hydrophobic residues, V37H, L89L, F98L, and W104H, are close to the methoxy group of mPEG, thereby raising an important question, how h15-2b specifically binds to PEG molecules with a terminal methoxy group. Therefore, we generated L89LD, F98LE, F98LY, W104HY, W104HE V37HS, and V37HT Fab variants, in which the hydrophobic side chains were replaced by the polar side chains to test whether these mutants can switch their binding specificity from mPEG to OH-PEG or SH-PEG. None of the h15-2b variants bound to SH-PEG while L89LD, F98LE, and W104HE variants exhibited impaired binding to mPEG and OH-PEG (Supplementary Fig. 4; Fig. 4a; Supplementary Table 1). By contrast, F98LY variant maintained similar mPEG-binding as compared to wild-type h15-2b but showed limited interaction with OH-PEG (Fig. 4a; Supplementary Table 1). Interestingly, mutating the hydrophobic W104H and V37H to a polar amino acid, tyrosine or serine, or threonine (W104HY, V37HS, and V37HT) enhanced mPEG-binding and mildly increased binding to OH-PEG, possibly by formation of hydrogen bonds to methoxy and hydroxyl groups on the mPEG and OH-PEG molecules, respectively (Fig. 4b). Taken together, Fig. 4 reveals that hydrophilic amino acids (L89LD, F98LE, and W104HE) might reduce binding to mPEG and OH-PEG. On the other hand, all h15-2b variants are unable to bind OH-PEG, except for W104HY, V37HS, and V37HT mutants, which mildly increase binding to OH-PEG, indicating that W104H and V37H is critical for specificity to mPEG (Supplementary Fig. 4; Supplementary Table 1).
Discussion
We report the first crystal structure of an antibody that specifically binds PEG with a terminal methoxy group. Aromatic amino acids in both framework and CDR regions stabilize the hook-shaped CR-like structure of the PEG chain via van der Waals and H-bond interactions. Hydrophobic residues, including V37H, L89L, F98L, and W104H, provide the methoxy-specificity of h15-2b, which are also highly conserved throughout antibodies to maintain the stability of the VH:VL interface32. Interestingly, V37HS and V37HT variants can partially cross-react with OH-PEG, indicating that V37H is mostly important for the selectivity of h15-2b toward methoxy-terminated PEG.
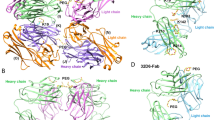
Note that although V37H, L89L, F98L, and W104H residues are responsible for the methoxy-specificity of h15-2b, these residues are highly conserved throughout anti-PEG and anti-mPEG antibodies (Supplementary Fig. 5). We found that h15-2b possesses shorter heavy chain complementarity-determining region 3 (HCDR3) but similar light chain CDR3 (LCDR3) as compared to 3.3, 2B5, and 6.3 (Supplementary Fig. 5). Structural comparison of the anti-PEG and anti-mPEG Fabs reveals that the h15-2b HCDR3 loop coupled with these conserved residues assist to form a tunnel-like structure for capturing the mPEG deep in the VH:VL interface (Supplementary Fig. 6a). By contrast, the bulky HCDR3 loops of these anti-PEG Fabs hinder the tunnel-like structure formation and provide large surface area to interact with the backbone of PEG (Supplementary Fig. 6b–d). Therefore, this might explain the unique methoxy-specificity of h15-2b.
The crystal structures of several backbone-specific anti-PEG antibodies in complex with PEG have been determined. Unlike 3.3, 2B5 and 6.3 anti-PEG backbone-specific antibodies that dimerize while interacting with PEG24,25, h15-2b binds mPEG as a monomer. Briefly, the PEG chain forms a planar or spiral S-shaped CR-like configuration while bridging two Fabs in their antigen-binding sites to form 3.3, 2B5, and 6.3 Fab homodimers (Supplementary Fig. 7). These planar symmetric CR-like structures of PEG are bound by backbone-specific anti-PEG antibodies via aromatic residues and coordinate with water molecules to form H-bond with the ether oxygen atoms of PEG24. By contrast, the mPEG chain is clamped by aromatic amino acids in the paratope surface area of h15-2b to form the first CR-like structure and then bent into the VH:VL interface to form a second CR-like structure (Supplementary Fig. 7). Indeed, CR is a cyclic version of linear PEG that exhibits a more stable configuration. Therefore, despite PEG polymer being highly flexible with variant conformations, the similar CR-like structures clamped by aromatic amino acids or the guanidinium moiety of arginine are observed in both backbone-specific anti-PEG antibodies and the methoxy-specific h15-2b anti-mPEG antibody.
mPEG is widely used in medical products to improve the solubility and circulation time of therapeutic proteins and nanoparticles. Therefore, both anti-PEG and anti-mPEG antibodies generated in patients might hinder the therapeutic efficacy and safety of PEGylated medicine. Although pre-existing anti-PEG antibodies are present in many naïve individuals, the incidence of pre-existing anti-mPEG antibodies remains unexplored. Most assays were developed for measuring anti-PEG antibodies in serum samples using direct ELISA for binding to immobilized mPEG18,21. However, these direct ELISA assays are unable to distinguish anti-PEG and anti-mPEG antibodies since coated mPEG could be detected by both. Alternatively, we hypothesize that anti-mPEG antibodies might be employed for competitive ELISA to measure pre-existing anti-mPEG antibodies22. Supposedly, serum samples containing pre-existing anti-mPEG but not anti-PEG antibodies can compete with mPEG-biotin captured by immobilized h15-2b antibodies to obtain a dose-response curve.
Engineered bispecific PEG-binding antibodies (PEG engagers) that simultaneously bind PEG molecules and tumor-associated antigens have also been developed for targeted delivery of mPEGylated nanomedicine in cancer therapies33,34,35,36,37,38. Intriguingly, the efficacy of PEG engager-directed mPEGylated nanomedicine is not compromised by pre-existing anti-PEG antibodies due to the high anti-PEG affinity of the PEG engagers35. Unlike chemical conjugation of targeting ligands with mPEGylated nanomedicine that require complicated covalent conjugation process, PEG engagers allow non-covalent mixing with any mPEGylated nanomedicine to reduce manufacturing complexity for targeted therapy. However, it has been reported that non-covalent antibody-payload complex dissociate in vivo after extended periods of time and presumably influence their effectiveness39. To further improve the in vivo long-term stability of PEG engager-directed nanomedicine, affinity improvement of anti-mPEG antibodies is required. In conclusion, the structural and functional analyses of h15-2b not only elucidate the mPEG-binding mechanism of anti-mPEG antibodies but also provide useful insights to assist in the rational design of high-affinity anti-mPEG antibodies. This might be accomplished by the combination of the W104HY, V37HS and V37HT mutations to create high-affinity h15-2b variants for the analytical study of pre-existing anti-mPEG antibodies and the development of efficient targeted mPEG-nanomedicine.
Methods
Production of humanized anti-mPEG and anti-PEG Fabs
The VL-Cκ and VH-CH1 DNA fragments of anti-mPEG h15-2b or anti-PEG 6.3 were linked by a composite internal ribosome entry site bicistronic expression element and inserted into the pLPCX plasmid containing a C-terminal polyhistidine-tag to generate pLPCX-anti-mPEG Fab-6xHis and pLPCX-anti-PEG Fab-6xHis plasmids. ExpiCHO-S cells were transfected with pLPCX-anti-mPEG Fab-6xHis and pLPCX-anti-PEG Fab-6xHis using ExpiFectamine CHO Transfection Kit (Thermo Fisher Scientific, San Jose, CA) according to the manufacturer’s instructions. Culture supernatant was harvested 10 days post-transfection by centrifugation at 1000 x g for 5 min and then filtered through a 0.45 μm filter. The polyhistidine-tagged Fab was purified on a HiTrap TALON crude column (Cytiva, Marlborough, MA). Protein concentrations were determined by the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, San Jose, CA).
Crystallization and data collection
Anti-mPEG h15-2b Fab was crystallized using the hanging-drop vapor diffusion method. The anti-mPEG Fab was buffer exchanged in 20 mmol L−1 Tris containing 100 mmol L−1 NaCl, pH 7.5, and concentrated to 21 mg mL−1. The complex crystals of the anti-mPEG Fab/mPEG were grown by mixing 1 μL protein solution (21 mg mL−1) with 1 μL reservoir solution using the sitting-drop vapor diffusion method at 20 °C. The anti-mPEG Fab/mPEG crystals were crystallized under a screen condition: 18% (w/v) PEG-6000, 1% (w/v) PEG-2000 methyl ether, 0.15 mol L−1 lithium sulfate monohydrate, and 0.1 mol L−1 citric acid (pH 3.5) at 20 °C (Sigma–Aldrich, St. Louis, MO). Snap-freezing the crystals with 20% glycerol (v/v) as a cryoprotectant for X-ray data collection at cryogenic temperatures (80 K). The diffraction data of anti-mPEG Fab/mPEG crystals were collected at the National Synchrotron Radiation Research Center (NSRRC, Taiwan) on beamline BL13B1 using a wavelength of 0.9732 Å with the ADSC QUANTUM 315r CCD detector (Area Detector Systems Corporation, Poway, CA). Data were indexed and scaled using HKL2000.
Structure determination and refinement
The crystal structure of anti-mPEG Fab in complex with mPEG was determined by the molecular replacement method of MOLREP using the structure of the anti-ErbB2 Fab2C4 (Protein Data Bank entry 1L7I) as a search model29. Extensive manual model building and refinement were performed using COOT40. The models were further refined with REFMAC41. The final model shows 0.48% outliers and 95.04% of residues in the most favored regions and 4.48% of residues in the additionally allowed regions of the Ramachandran diagram. The atomic coordinates and structure factors of h15-2b Fab/mPEG were deposited in the Protein Data Bank as entry 7Y0G. PyMol was used to generate figures of structures.
Generation of h15-2b Fab variants by site-directed mutagenesis
The wild-type light chain and heavy chain with a C-terminal polyhistidine-tag of h15-2b Fab were cloned into a dual-expression plasmid containing the pelB and the stII signal peptides for periplasm expression in Escherichia coli. The parental h15-2b Fab plasmid was employed as a template to amplify h15-2b VH and VL mutant fragments by site-directed mutagenesis using overlap extension PCR30. The parental plasmids were linearized by digesting with BsiwI/NheI or NcoI/RsrII (New England Biolabs, Ipswich, MA) to remove original VH or VL fragments, respectively. h15-2b mutant amplicons were cloned into the linearized vector using Gibson Assembly cloning. Each of the h15-2b Fab variants was transformed into E. coli C43 (DE3). The resultant recombinant strains were culture in 2xYT Broth at 37 °C and induced with 1 mmol L−1 of isopropyl β-D-1-thiogalactopyranoside (IPTG) (Sigma–Aldrich, St. Louis, MO) when an OD600 of 0.5 was reached. The cells were incubated at 30 °C for 20 h after IPTG induction. The cells were collected by centrifugation at 3000 x g for 15 min at 4 °C, and the pellet was resuspended in lysis buffer (100 mmol L−1 Tris, 300 mmol L−1 NaCl, 1 mmol L−1 phenylmethanesulfonyl fluoride, pH 8.0) (Sigma–Aldrich, St. Louis, MO). After sonication, the clear lysate was harvested by centrifugation at 12,000 x g for 15 min at 4 °C and filtered through a 0.45 μm filter. Each polyhistidine-tagged h15-2b mutant Fabs was purified on a HiTrap TALON crude column (Cytiva, Marlborough, MA) and buffer exchanged in PBS buffer for further characterization.
Antibody ELISA
Maxisorp 96-well microplates (Thermo Fisher Scientific, San Jose, CA) were coated with 0.5 µg per well of mPEG5K-NH2 or OH-PEG5K-NH2 (Nanocs, New York, NY) or SH-PEG3.5K-NH2 (Sigma–Aldrich, St. Louis, MO) or recombinant GFP (Cell Biolabs, Inc., San Diego, CA) in 50 µL 100 mmol L−1 NaHCO3/Na2CO3 coating buffer (pH 8.0) for 3 h at 37 °C and then blocked with 200 µL of 5% (wt/vol) skim milk in PBS at 4 °C overnight. Graded concentrations of purified h15-2b Fab variants or 6.3 Fab or anti-GFP Fab in 50 µL 2% (wt/vol) skim milk was added to the plates at RT for 1 h. The plates were washed with PBS three times. HRP-conjugated goat anti-human F(ab’)2 fragment specific antibodies (1 µg mL−1) (Jackson Immuno Research Laboratories, West Grove, PA, catalog number: 109-035-097) in 50 µL 2% (wt/vol) skim milk was added for 30 min at room temperature. The plates were washed with PBS three times, and bound peroxidase activity was measured by adding 150 µL per well of ABTS substrate solution (0.4 mg mL−1 2,2’-azino-di (3-ethylbenzthiazoline-6-sulfonic acid) (Sigma–Aldrich, St. Louis, MO), 0.003% H2O2, 100 mmol L−1 phosphate citrate, pH 4.0) for 30 min at room temperature. The absorbance (405 nm) was measured in a SpectraMax ABS Plus microplate reader (Molecular Device, Menlo Park, CA). GraphPad Prism 6 was used to analyze ELISA data. The anti-GFP Fab was kindly provided by Dr. Kurt Yun Mou (Academia Sinica, Taipei, Taiwan). Note that both Maxisorp (Thermo Fisher Scientific, San Jose, CA) and Well-Coated™ Amine Binding microplates (G-Biosciences, St. Louis, MO) are widely used for immobilization of amine group containing antigens. Comparison of these microplates for anti-PEG ELISA revealed that there is no difference between them (Supplementary Fig. 8). Therefore, the Maxisorp microplates were used for all ELISA experiments in this study.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are available from the corresponding author on reasonable request. The Protein Data Bank data of h15-2b Fab/mPEG crystal structure can be found in Supplementary Data 1. The atomic coordinates and structure factors of h15-2b Fab/mPEG were deposited in the Protein Data Bank as entry 7Y0G.
References
Harris, J. M. & Chess, R. B. Effect of pegylation on pharmaceuticals. Nat. Rev. Drug Discov. 2, 214–221 (2003).
Alconcel, S. N. S., Baas, A. S. & Maynard, H. D. FDA-approved poly(ethylene glycol)–protein conjugate drugs. Polym. Chem. 2, 1442–1448 (2011).
Swierczewska, M., Lee, K. C. & Lee, S. What is the future of PEGylated therapies? Expert. Opin. Emerg. Drugs 20, 531–536 (2015).
Chey, W. D. et al. Naloxegol for opioid-induced constipation in patients with noncancer pain. N. Engl. J. Med. 370, 2387–2396 (2014).
Jahnson, S., Sandblom, D. & Holmäng, S. A randomized trial comparing 2 doses of polidocanol sclerotherapy for hydrocele or spermatocele. J. Urol. 186, 1319–1323 (2011).
Heathcote, E. J. et al. Peginterferon alfa-2a in patients with chronic hepatitis C and cirrhosis. N. Engl. J. Med. 343, 1673–1680 (2000).
Manns, M. P. et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358, 958–965 (2001).
Reddy, K. R. et al. Efficacy and safety of pegylated (40-kd) interferon alpha-2a compared with interferon alpha-2a in noncirrhotic patients with chronic hepatitis C. Hepatology 33, 433–438 (2001).
Levin, N. W. et al. Intravenous methoxy polyethylene glycol-epoetin beta for haemoglobin control in patients with chronic kidney disease who are on dialysis: a randomised non-inferiority trial (MAXIMA). Lancet 370, 1415–1421 (2007).
Safra, T. et al. Pegylated liposomal doxorubicin (doxil): reduced clinical cardiotoxicity in patients reaching or exceeding cumulative doses of 500 mg/m2. Ann. Oncol. 11, 1029–1033 (2000).
Rafiyath, S. M. et al. Comparison of safety and toxicity of liposomal doxorubicin vs. conventional anthracyclines: a meta-analysis. Exp. Hematol. Oncol. 1, 10 (2012).
Knop, K., Hoogenboom, R., Fischer, D. & Schubert, U. S. Poly(ethylene glycol) in drug delivery: pros and cons as well as potential alternatives. Angew. Chem. Int. Ed. 49, 6288–6308 (2010).
Chang, T. C. et al. Both IgM and IgG Antibodies Against Polyethylene Glycol Can Alter the Biological Activity of Methoxy Polyethylene Glycol-Epoetin Beta in Mice. Pharmaceutics 12 (2019).
Hsieh, Y. C. et al. Pre-existing anti-polyethylene glycol antibody reduces the therapeutic efficacy and pharmacokinetics of PEGylated liposomes. Theranostics 8, 3164–3175 (2018).
Armstrong, J. K. et al. Antibody against poly(ethylene glycol) adversely affects PEG-asparaginase therapy in acute lymphoblastic leukemia patients. Cancer 110, 103–111 (2007).
Dams, E. T. et al. Accelerated blood clearance and altered biodistribution of repeated injections of sterically stabilized liposomes. J. Pharmacol. Exp. Ther. 292, 1071–1079 (2000).
Chen, E. et al. Premature Drug Release from Polyethylene Glycol (PEG)-Coated Liposomal Doxorubicin via Formation of the Membrane Attack Complex. ACS Nano 14, 7808–7822 (2020).
Chen, B. M. et al. Measurement of Pre-Existing IgG and IgM Antibodies against Polyethylene Glycol in Healthy Individuals. Anal. Chem. 88, 10661–10666 (2016).
Yang, Q. et al. Analysis of Pre-existing IgG and IgM Antibodies against Polyethylene Glycol (PEG) in the General Population. Anal. Chem. 88, 11804–11812 (2016).
McSweeney, M. D., Mohan, M., Commins, S. P. & Lai, S. K. Anaphylaxis to Pfizer/BioNTech mRNA COVID-19 Vaccine in a Patient With Clinically Confirmed PEG Allergy. Front. Allergy 2 (2021).
Chen, B.-M., Cheng, T.-L. & Roffler, S. R. Polyethylene Glycol Immunogenicity: Theoretical, Clinical, and Practical Aspects of Anti-Polyethylene Glycol Antibodies. ACS Nano 15, 14022–14048 (2021).
Chuang, K.-H. et al. Development of an Anti-Methoxy Poly(ethylene glycol) (α-mPEG) Cell-Based Capture System to Measure mPEG and mPEGylated Molecules. Macromolecules 47, 6880–6888 (2014).
Lin, W.-W. et al. Optimization of an Anti-poly(ethylene glycol) (anti-PEG) Cell-Based Capture System To Quantify PEG and PEGylated Molecules. Anal. Chem. 88, 12371–12379 (2016).
Lee, C. C. et al. Structural basis of polyethylene glycol recognition by antibody. J. Biomed. Sci. 27, 12 (2020).
Huckaby, J. T. et al. Structure of an anti-PEG antibody reveals an open ring that captures highly flexible PEG polymers. Commun. Chem. 3, 124 (2020).
Sherman, M. R., Williams, L. D., Sobczyk, M. A., Michaels, S. J. & Saifer, M. G. P. Role of the Methoxy Group in Immune Responses to mPEG-Protein Conjugates. Bioconjug. Chem. 23, 485–499 (2012).
Saifer, M. G., Williams, L. D., Sobczyk, M. A., Michaels, S. J. & Sherman, M. R. Selectivity of binding of PEGs and PEG-like oligomers to anti-PEG antibodies induced by methoxyPEG-proteins. Mol. Immunol. 57, 236–246 (2014).
Shimizu, T. et al. A hydroxyl PEG version of PEGylated liposomes and its impact on anti-PEG IgM induction and on the accelerated clearance of PEGylated liposomes. Eur. J. Pharm. Biopharm. 127, 142–149 (2018).
Vajdos, F. F. et al. Comprehensive functional maps of the antigen-binding site of an anti-ErbB2 antibody obtained with shotgun scanning mutagenesis. J. Mol. Biol. 320, 415–428 (2002).
Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K. & Pease, L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 (1989).
Lee, C. C. et al. Crowning proteins: modulating the protein surface properties using crown ethers. Angew. Chem. Int. Ed. Engl. 53, 13054–13058 (2014).
Herold, E. M. et al. Determinants of the assembly and function of antibody variable domains. Sci. Rep. 7, 12276 (2017).
Kao, C.-H. et al. One-step mixing with humanized anti-mPEG bispecific antibody enhances tumor accumulation and therapeutic efficacy of mPEGylated nanoparticles. Biomaterials 35, 9930–9940 (2014).
Tung, H. Y. et al. Selective Delivery of PEGylated Compounds to Tumor Cells by Anti-PEG Hybrid Antibodies. Mol. Cancer Ther. 14, 1317–1326 (2015).
Su, Y.-C. et al. Conditional internalization of PEGylated nanomedicines by PEG engagers for triple negative breast cancer therapy. Nat. Commun. 8, 15507 (2017).
Cheng, Y. A. et al. Enhanced drug internalization and therapeutic efficacy of PEGylated nanoparticles by one-step formulation with anti-mPEG bispecific antibody in intrinsic drug-resistant breast cancer. Biomater. Sci. 7, 3404–3417 (2019).
Chen, I. J. et al. Bispecific antibody (HER2 × mPEG) enhances anti-cancer effects by precise targeting and accumulation of mPEGylated liposomes. Acta Biomater. 111, 386–397 (2020).
Ho, K.-W. et al. Double attack strategy for leukemia using a pre-targeting bispecific antibody (CD20 Ab-mPEG scFv) and actively attracting PEGylated liposomal doxorubicin to enhance anti-tumor activity. J. Nanobiotechnology 19, 16 (2021).
Dengl, S. et al. Hapten-directed spontaneous disulfide shuffling: a universal technology for site-directed covalent coupling of payloads to antibodies. FASEB J. 29, 1763–1779 (2015).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 (1997).
Acknowledgements
This work is financially supported by the Ministry of Science and Technology (MOST) of Taiwan under grant number MOST110-2113-M-A49-026-MY3 to C.-Y.C. and through the Young Scholar Fellowship (Columbus) Program under grant number MOST108-2636-B-009-003, MOST109-2636-B-009-003, MOST110-2636-B-009-010, and MOST111-2636-B-A49-005 to Y.-C.S. This work is also supported in part by the Kaohsiung Medical University Research Center Grant (NYCUKMU-111-I005) and the “Center for Intelligent Drug Systems and Smart Bio-devices (IDS2B)” from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan. The E. coli C43 (DE3) and anti-GFP Fab were kindly offered by Dr. Kurt Yun Mou (Academia Sinica, Taipei, Taiwan). The authors also thank the technical services provided by the Synchrotron Radiation Protein Crystallography Facility of the National Core Facility Program for Biotechnology, MOST, and the National Synchrotron Radiation Research Center (NSRRC), a national user facility supported by MOST, Taiwan.
Author information
Authors and Affiliations
Contributions
M.-T.T.N., Y.-C. Shih, C.-Y.H. and M.-H.L. performed the experiments. M.-T.T.N., Y.-C. Shih, E.-C.L., Y.-J.J., W.-W.L. and S.R.R. analyzed and interpreted the data. M.-T.T.N., M.-H.L., C.-Y.C. and Y.-C. Su analyzed protein crystallization data. T.-L.C., C.-Y.C., S.R.R. and Y.-C. Su wrote the manuscript. C.-Y.C. and Y.-C. Su designed and supervised the project. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Chemistry thanks Nicolas Bertrand, Alastair Lawson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nguyen, MT.T., Shih, YC., Lin, MH. et al. Structural determination of an antibody that specifically recognizes polyethylene glycol with a terminal methoxy group. Commun Chem 5, 88 (2022). https://doi.org/10.1038/s42004-022-00709-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42004-022-00709-0
- Springer Nature Limited